Abstract
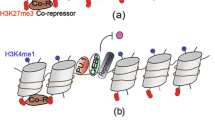
Macrophages provide a critical systemic network cells of the innate immune system. Emerging data suggest that in addition, they have important tissue-specific functions that range from clearance of surfactant from the lungs to neuronal pruning and establishment of gut homeostasis. The differentiation and tissue-specific activation of macrophages require precise regulation of gene expression, a process governed by epigenetic mechanisms such as DNA methylation, histone modification and chromatin structure. We argue that epigenetic regulation of macrophages is determined by lineage- and tissue-specific transcription factors controlled by the built-in programming of myeloid development in combination with signaling from the tissue environment. Perturbation of epigenetic mechanisms of tissue macrophage identity can affect normal macrophage tissue function and contribute to pathologies ranging from obesity and autoimmunity to neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
24 October 2016
In the version of this article initially published, the citation of Figure 3 in the second paragraph of the third subsection (Chromatin: the nexus of phenotype and the environment) is incorrect. That should cite Figure 2, as follows: "From an evolutionary perspective, two extreme models of how such complexity might be generated and regulated can be envisaged95 (Fig. 2)." The error has been corrected in the HTML and PDF versions of the article.
References
Ueno, M. & Yamashita, T. Bidirectional tuning of microglia in the developing brain: from neurogenesis to neural circuit formation. Curr. Opin. Neurobiol. 27, 8–15 (2014).
Fulde, M. & Hornef, M.W. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol. Rev. 260, 21–34 (2014).
Wynn, T.A., Chawla, A. & Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Gordon, S., Plüddemann, A. & Martinez Estrada, F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 262, 36–55 (2014).
Sieweke, M.H. & Allen, J.E. Beyond stem cells: self-renewal of differentiated macrophages. Science 342, 1242974–1242974 (2013).
Mucenski, M.L. et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–689 (1991).
Perdiguero, E.G. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Hoeffel, G. et al. C-Myb+ erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Orkin, S.H. & Zon, L.I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Bain, C.C. et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15, 929–937 (2014).
Molawi, K. et al. Progressive replacement of embryo-derived cardiac macrophages with age. J. Exp. Med. 211, 2151–2158 (2014).
Varol, C. et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204, 171–180 (2007).
Hashimoto, D., Miller, J. & Merad, M. Dendritic cell and macrophage heterogeneity in vivo. Immunity 35, 323–335 (2011).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Bruttger, J. et al. Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity 43, 92–106 (2015).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Malissen, B., Tamoutounour, S. & Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 14, 417–428 (2014).
Zigmond, E. & Jung, S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 34, 162–168 (2013).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Gordon, S. The macrophage: past, present and future. Eur. J. Immunol. 37, S9–S17 (2007).
Chow, A. et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 208, 261–271 (2011).
Muller, P.A. et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158, 300–313 (2014).
Niess, J.H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Parkhurst, C.N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013).
Berg vom, J. et al. Inhibition of IL-12/IL-23 signaling reduces Alzheimer's disease–like pathology and cognitive decline. Nat. Med. 18, 1812–1819 (2012).
Yamasaki, R. et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med. 211, 1533–1549 (2014).
Chiu, I.M. et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013).
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Davalos, D. et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758 (2005).
Nimmerjahn, A., Kirchhoff, F. & Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005).
Tremblay, M.-È., Lowery, R.L. & Majewska, A.K. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 8, e1000527 (2010).
Schafer, D.P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012).
Paolicelli, R.C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Squarzoni, P. et al. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8, 1271–1279 (2014).
Prinz, M. & Priller, J. Microglia and brain macrophagesin the molecular age: from origin toneuropsychiatric disease. Nat. Rev. Neurosci. 5, 300–312 (2014).
Rademakers, R. et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 44, 200–205 (2012).
Poliani, P.L. et al. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125, 2161–2170 (2015).
Guerreiro, R. & Hardy, J. TREM2 and neurodegenerative disease. N. Engl. J. Med. 369, 1569–1570 (2013).
Goldmann, T. et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16, 1618–1626 (2013).
Bogunovic, M. et al. Origin of the lamina propria dendritic cell network. Immunity 31, 513–525 (2009).
Varol, C. et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 31, 502–512 (2009).
Ginhoux, F. & Jung, S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014).
Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Weber, B., Saurer, L., Schenk, M., Dickgreber, N. & Mueller, C. CX3CR1 defines functionally distinct intestinal mononuclear phagocyte subsets which maintain their respective functions during homeostatic and inflammatory conditions. Eur. J. Immunol. 41, 773–779 (2011).
Zigmond, E. et al. Ly6Chi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090 (2012).
Rubtsov, Y.P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Shouval, D.S. et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not il-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Mortha, A. et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343, 1249288–1249288 (2014).
Seno, H. et al. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc. Natl. Acad. Sci. USA 106, 256–261 (2009).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Schneider, C. et al. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 10, e1004053 (2014).
Suzuki, T. et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J. Exp. Med. 205, 2703–2710 (2008).
Nakamura, A. et al. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J. Exp. Med. 210, 2191–2204 (2013).
Happle, C. et al. Pulmonary transplantation of macrophage progenitors as effective and long-lasting therapy for hereditary pulmonary alveolar proteinosis. Sci. Transl. Med. 6, 250ra113 (2014).
Suzuki, T. et al. Pulmonary macrophage transplantation therapy. Nature 514, 450–454 (2014).
Mebius, R.E. & Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 5, 606–616 (2005).
Miyake, Y. et al. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell–associated antigens. J. Clin. Invest. 117, 2268–2278 (2007).
A-Gonzalez, N. et al. The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat. Immunol. 14, 831–839 (2013).
Ganz, T. Macrophages and systemic iron homeostasis. J. Innate Immun. 4, 446–453 (2012).
Kohyama, M. et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457, 318–321 (2009).
Glocker, E.-O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Farh, K.K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Gautier, E.L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Jaitin, D.A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Jojic, V. et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14, 633–643 (2013).
Miller, J.C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Cipolletta, D., Kolodin, D., Benoist, C. & Mathis, D. Tissular Tregs: a unique population of adipose-tissue-resident Foxp3+CD4+ T cells that impacts organismal metabolism. Semin. Immunol. 23, 431–437 (2011).
Haldar, M. et al. Heme-mediated Spi-c induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Cohen, M. et al. Chronic exposure to TGFβ1 regulates myeloid cell inflammatory response in an IRF7-dependent manner. EMBO J. 33, 2906–2921 (2014).
Davalos, D. et al. Fibrinogen-induced perivascular microglial clustering is required for the development of axonal damage in neuroinflammation. Nat. Commun. 3, 1227 (2012).
Erny, D. et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015).
Thorburn, A.N., Macia, L. & Mackay, C.R. Diet, metabolites, and “Western-lifestyle” inflammatory diseases. Immunity 40, 833–842 (2014).
Ghosn, E.E.B. et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl. Acad. Sci. USA 107, 2568–2573 (2010).
Gautier, E.L. et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211, 1525–1531 (2014).
Rosas, M. et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648 (2014).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 (2014).
Capucha, T. et al. Distinct murine mucosal langerhans cell subsets develop from pre-dendritic cells and monocytes. Immunity 43, 369–381 (2015).
Garber, M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ostuni, R. & Natoli, G. Lineages, cell types and functional states: a genomic view. Curr. Opin. Cell Biol. 25, 759–764 (2013).
Winter, D.R. & Amit, I. The role of chromatin dynamics in immune cell development. Immunol. Rev. 261, 9–22 (2014).
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Zhou, V.W., Goren, A. & Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 (2011).
Lawrence, T. & Natoli, G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 (2011).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Schultze, J.L., Freeman, T., Hume, D.A. & Latz, E. A transcriptional perspective on human macrophage biology. Semin. Immunol. 27, 44–50 (2015).
Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Lee, T.I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Nord, A.S. et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013).
Zhang, J.A., Mortazavi, A., Williams, B.A., Wold, B.J. & Rothenberg, E.V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482 (2012).
Bauer, D.E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013).
Heinz, S. & Glass, C.K. Roles of lineage-determining transcription factors in establishing open chromatin: lessons from high-throughput studies. Curr. Top. Microbiol. Immunol. 356, 1–15 (2012).
Bossard, P. & Zaret, K.S. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125, 4909–4917 (1998).
Cirillo, L.A. & Zaret, K.S. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961–969 (1999).
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008).
Cirillo, L.A. et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002).
Bornstein, C. et al. A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin states. Mol. Cell 56, 749–762 (2014).
Graf, T. & Enver, T. Forcing cells to change lineages. Nature 462, 587–594 (2009).
Feng, R. et al. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 105, 6057–6062 (2008).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Acknowledgements
Supported by the European Research Council (309788 for the I.A. laboratory; 340345 for the S.J. laboratory), the I-CORE for chromatin and RNA regulation (I.A. laboratory), the Israel Science Foundation (703/15 for the I.A. laboratory; 887/11 for the S.J. laboratory), the BLUEPRINT FP7 consortium (I.A. laboratory), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine (I.A. laboratory), Minerva Stiftung (I.A. laboratory), The Azrieli Foundation (D.R.W.), the European Molecular Biology Organization (ALT766-2014 for D.R.W.) and the European Commission FP7 (Marie Curie Actions, EMBOCOFUND2012, GA-2012-600394 for D.R.W.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Amit, I., Winter, D. & Jung, S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol 17, 18–25 (2016). https://doi.org/10.1038/ni.3325
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3325
This article is cited by
-
Myeloid cell-targeted therapies for solid tumours
Nature Reviews Immunology (2023)
-
Integrated proteomic and transcriptomic landscape of macrophages in mouse tissues
Nature Communications (2022)
-
Macrophages as tools and targets in cancer therapy
Nature Reviews Drug Discovery (2022)
-
Inhibition of EZH2 prevents acute respiratory distress syndrome (ARDS)-associated pulmonary fibrosis by regulating the macrophage polarization phenotype
Respiratory Research (2021)
-
Immunological memory in rheumatic inflammation — a roadblock to tolerance induction
Nature Reviews Rheumatology (2021)