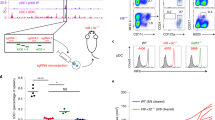
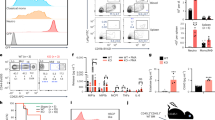
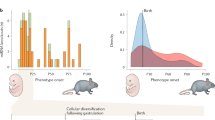
Abstract
To investigate if the microRNA (miRNA) pathway is required for dendritic cell (DC) development, we assessed the effect of ablating Drosha and Dicer, the two enzymes central to miRNA biogenesis. We found that while Dicer deficiency had some effect, Drosha deficiency completely halted DC development and halted myelopoiesis more generally. This indicated that while the miRNA pathway did have a role, it was a non-miRNA function of Drosha that was particularly critical. Drosha repressed the expression of two mRNAs encoding inhibitors of myelopoiesis in early hematopoietic progenitors. We found that Drosha directly cleaved stem-loop structure within these mRNAs and that this mRNA degradation was necessary for myelopoiesis. We have therefore identified a mechanism that regulates the development of DCs and other myeloid cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Kondo, M., Weissman, I.L. & Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672 (1997).
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000).
Naik, S.H. et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007).
Onai, N. et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007).
Corcoran, L. et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J. Immunol. 170, 4926–4932 (2003).
D'Amico, A. & Wu, L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J. Exp. Med. 198, 293–303 (2003).
Onai, N., Obata-Onai, A., Tussiwand, R., Lanzavecchia, A. & Manz, M.G. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J. Exp. Med. 203, 227–238 (2006).
Carotta, S. et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32, 628–641 (2010).
Belz, G.T. & Nutt, S.L. Transcriptional programming of the dendritic cell network. Nat. Rev. Immunol. 12, 101–113 (2012).
Chong, M.M. et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24, 1951–1960 (2010).
Koralov, S.B. et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874 (2008).
Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Grishok, A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001).
Hutvágner, G. et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001).
Ketting, R.F. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001).
Li, H.S., Greeley, N., Sugimoto, N., Liu, Y.J. & Watowich, S.S. miR-22 controls Irf8 mRNA abundance and murine dendritic cell development. PLoS ONE 7, e52341 (2012).
Mildner, A. et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood 121, 1016–1027 (2013).
Alemdehy, M.F. et al. Dicer1 deletion in myeloid-committed progenitors causes neutrophil dysplasia and blocks macrophage/dendritic cell development in mice. Blood 119, 4723–4730 (2012).
Guo, S. et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA 107, 14229–14234 (2010).
Kuipers, H., Schnorfeil, F.M. & Brocker, T. Differentially expressed microRNAs regulate plasmacytoid vs. conventional dendritic cell development. Mol. Immunol. 48, 333–340 (2010).
Gantier, M.P. et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39, 5692–5703 (2011).
Merad, M. & Manz, M.G. Dendritic cell homeostasis. Blood 113, 3418–3427 (2009).
Brasel, K., De Smedt, T., Smith, J.L. & Maliszewski, C.R. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood 96, 3029–3039 (2000).
Han, J. et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75–84 (2009).
Karginov, F.V. et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell 38, 781–788 (2010).
Kirigin, F.F. et al. Dynamic microRNA gene transcription and processing during T cell development. J. Immunol. 188, 3257–3267 (2012).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Basu, S. et al. “Emergency” granulopoiesis in G-CSF-deficient mice in response to Candida albicans infection. Blood 95, 3725–3733 (2000).
Manel, N. et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467, 214–217 (2010).
Johanson, T.M., Lew, A.M. & Chong, M.M. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 3, 130144 (2013).
Knuckles, P. et al. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 15, 962–969 (2012).
Cheloufi, S., Dos Santos, C.O., Chong, M.M. & Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 (2010).
Johanson, T.M. et al. A microRNA expression atlas of mouse dendritic cell development. Immunol. Cell Biol. 93, 480–485 (2015).
Shenoy, A. & Blelloch, R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS ONE 4, e6971 (2009).
Chong, M.M., Rasmussen, J.P., Rudensky, A.Y. & Littman, D.R. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205, 2005–2017 (2008).
Teta, M. et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development 139, 1405–1416 (2012).
Zhdanova, O. et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 80, 719–730 (2011).
Gromak, N. et al. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 5, 1499–1510 (2013).
Davis, B.N. & Hata, A. Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun. Signal. 7, 18 (2009).
Newman, M.A., Thomson, J.M. & Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 (2008).
Naik, S.H. et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 7, 663–671 (2006).
Ding, Y. et al. FLT3-ligand treatment of humanized mice results in the generation of large numbers of CD141+ and CD1c+ dendritic cells in vivo. J. Immunol. 192, 1982–1989 (2014).
Lieschke, G.J. et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 (1994).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Ritchie, M.E. et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 7, 261 (2006).
Yang, Y.H. et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15 (2002).
Pear, W.S., Nolan, G.P., Scott, M.L. & Baltimore, D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90, 8392–8396 (1993).
Acknowledgements
We thank C. Yates for animal husbandry. Supported by the National Health and Medical Research Council, Australia (637338, 1004541, 1042211 and 1079586 to M.M.W.C.; 1037321, 1043414 and 1080321 to A.M.L.), the Juvenile Diabetes Research Foundation (5-2011-100 to M.M.W.C.), the Diabetes Australia Research Trust (Y13G-CHOM to M.M.W.C.), the Australian Research Council (M.M.W.C.) and the Victorian State Government Operational Infrastructure Support and Australian National Health and Medical Research Council Research Institute Infrastructure Support Scheme.
Author information
Authors and Affiliations
Contributions
T.M.J. performed most of the experiments with help from A.A.K., J.H.C.Y. and Y.Z.; M.C. and A.K. performed the bioinformatics analyses; and T.M.J., A.M.L., Y.Z. and M.M.W.C. designed the study, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Residual splenic DCs in Drosha-deficient chimeras have escaped deletion.
The residual splenic DCs in Tamoxifen-treated Droshafl/fl CreER chimeras were sorted by FACS, then Drosha mRNA levels were measured by quantitative RT-PCR.
Supplementary Figure 2 The accumulation of LSKs and CMPs caused by Drosha deficiency is cell intrinsic.
Comparison of LSK and CMP populations between the CD45.2 experimental and CD45.1 wild type supporting component in the bone marrow of chimeric mice following Tamoxifen treatment. The data shown is the mean +/- S.D. of twelve (control, Droshafl/fl CreER) or nine (Dicerfl/fl CreER) animals for each genotype from three independent experiments.
Supplementary Figure 3 Drosha ablation and miRNA expression are equivalent in LSK-, CMP-, CLP- and CDP-derived cultures.
Quantitative RT-PCR or Taqman-based quantitative RT-PCR determined levels of Drosha mRNA or a panel of mature miRNAs, respectively. The data shown is the mean +/- S.D. of one or two (LSK culture Drosha levels) independent experiments.
Supplementary Figure 4 Identification of cleavage sites within Myl9 mRNA and Todr1 mRNA.
(a) Schematic of 5’P capture experiment. PolyA transcripts were purified from total RNA isolated from lineage-depleted bone marrow cells. An adaptor was ligated specifically to transcripts exhibiting a 5’ monophosphate (a mark of endonucleolytic cleavage) and reverse transcribed. Tiled PCRs were then performed using a forward primer in the adaptor and reverse primers within Myl9 and Todr1 mRNA. Thus, PCR products could only be obtained if Myl9 and Todr1 contained a 5’ phosphate. PCR products were Sanger sequenced. The junction between the adaptor and mRNA denoted the original cleavage site. (b) Location of cleavage sites detected in Myl9 and Todr1 mRNA. Blue and red text denote stem or loop structure, respectively.
Supplementary Figure 5 Mutagenesis of the Todr1 stem-loop to disrupt base pairing.
Site-directed mutagenesis was used to introduce mismatch mutations into the sequence of the 5’ arm of the putative stem-loop structure within Todr1 mRNA.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1 and 2. (PDF 1482 kb)
Rights and permissions
About this article
Cite this article
Johanson, T., Keown, A., Cmero, M. et al. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat Immunol 16, 1134–1141 (2015). https://doi.org/10.1038/ni.3293
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3293
This article is cited by
-
Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo
Cellular and Molecular Life Sciences (2019)
-
RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding
Nature Structural & Molecular Biology (2017)
-
IL4 and IL21 cooperate to induce the high Bcl6 protein level required for germinal center formation
Immunology & Cell Biology (2017)
-
Cellular and molecular regulation of innate inflammatory responses
Cellular & Molecular Immunology (2016)
-
Drosha cuts the tethers of myelopoiesis
Nature Immunology (2015)