Abstract
The receptor NLRP3 is involved in the formation of the NLRP3 inflammasome that activates caspase-1 and mediates the release of interleukin 1β (IL-1β) and IL-18. Whether NLRP3 can shape immunological function independently of inflammasomes is unclear. We found that NLRP3 expression in CD4+ T cells specifically supported a T helper type 2 (TH2) transcriptional program in a cell-intrinsic manner. NLRP3, but not the inflammasome adaptor ASC or caspase-1, positively regulated a TH2 program. In TH2 cells, NLRP3 bound the Il4 promoter and transactivated it in conjunction with the transcription factor IRF4. Nlrp3-deficient TH2 cells supported melanoma tumor growth in an IL-4-dependent manner and also promoted asthma-like symptoms. Our results demonstrate the ability of NLRP3 to act as a key transcription factor in TH2 differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
12 October 2015
In the version of this article initially published, reference 44 (Farrell, G.A. Responding to aggression; the role of significant others for student psychiatric nurses, an ethnographic study. Nurse Educ. Today 9, 335–340 (1989)) was incorrect. The correct reference is as follows: Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012). The error has been corrected in the HTML and PDF versions of the article.
01 December 2015
Nat. Immunol. 16, 859–870 (2015); published online 22 June 2015; corrected after print 12 October 2015 In the version of this article initially published, reference 44 (Farrell, G.A. Responding to aggression; the role of significant others for student psychiatric nurses, an ethnographic study. NurseEduc.
References
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Anderson, O.A., Finkelstein, A. & Shima, D.T. A2E induces IL-1ss production in retinal pigment epithelial cells via the NLRP3 inflammasome. PLoS ONE 8, e67263 (2013).
Feron, V.J., Woutersen, R.A., van Garderen-Hoetmer, A. & Dreef-van der Meulen, H.C. Upper respiratory tract tumors in Cpb:WU (Wistar random) rats. Environ. Health Perspect. 85, 305–315 (1990).
Doitsh, G. et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 (2014).
Wang, W. et al. Inflammasome-independent NLRP3 augments TGF-β signaling in kidney epithelium. J. Immunol. 190, 1239–1249 (2013).
Hou, J., Schindler, U., Henzel, W.J., Wong, S.C. & McKnight, S.L. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity 2, 321–329 (1995).
Végran, F. et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat. Immunol. 15, 758–766 (2014).
Misawa, T. et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 (2013).
Bindea, G., Galon, J. & Mlecnik, B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663 (2013).
Rengarajan, J. et al. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195, 1003–1012 (2002).
Howard, M. et al. Identification of a T cell-derived B cell growth factor distinct from interleukin 2. J. Exp. Med. 155, 914–923 (1982).
Bruhn, K.W., Nelms, K., Boulay, J.L., Paul, W.E. & Lenardo, M.J. Molecular dissection of the mouse interleukin-4 promoter. Proc. Natl. Acad. Sci. USA 90, 9707–9711 (1993).
Kumar, R.K., Herbert, C. & Foster, P.S. The “classical” ovalbumin challenge model of asthma in mice. Curr. Drug Targets 9, 485–494 (2008).
Verma, C. et al. Abnormal T regulatory cells (Tregs: FOXP3+, CTLA-4+), myeloid-derived suppressor cells (MDSCs: monocytic, granulocytic) and polarised T helper cell profiles (Th1, Th2, Th17) in women with large and locally advanced breast cancers undergoing neoadjuvant chemotherapy (NAC) and surgery: failure of abolition of abnormal treg profile with treatment and correlation of treg levels with pathological response to NAC. J. Transl. Med. 11, 16 (2013).
Cândido, E.B. et al. Immune response evaluation through determination of type 1, type 2, and type 17 patterns in patients with epithelial ovarian cancer. Reprod. Sci. 20, 828–837 (2013).
Kobayashi, M., Kobayashi, H., Pollard, R.B. & Suzuki, F. A pathogenic role of Th2 cells and their cytokine products on the pulmonary metastasis of murine B16 melanoma. J. Immunol. 160, 5869–5873 (1998).
Hallett, M.A., Venmar, K.T. & Fingleton, B. Cytokine stimulation of epithelial cancer cells: the similar and divergent functions of IL-4 and IL-13. Cancer Res. 72, 6338–6343 (2012).
Powrie, F., Menon, S. & Coffman, R.L. Interleukin-4 and interleukin-10 synergize to inhibit cell-mediated immunity in vivo. Eur. J. Immunol. 23, 3043–3049 (1993).
Devaiah, B.N. & Singer, D.S. CIITA and Its Dual Roles in MHC Gene Transcription. Front. Immunol. 4, 476 (2013).
Cressman, D.E., O'Connor, W.J., Greer, S.F., Zhu, X.S. & Ting, J.P. Mechanisms of nuclear import and export that control the subcellular localization of class II transactivator. J. Immunol. 167, 3626–3634 (2001).
Grumont, R.J. & Gerondakis, S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor kappaB. J. Exp. Med. 191, 1281–1292 (2000).
Grossman, A. et al. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23-p25. Genomics 37, 229–233 (1996).
Staudt, V. et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202 (2010).
Bollig, N. et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc. Natl. Acad. Sci. USA 109, 8664–8669 (2012).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Lohoff, M. et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. USA 99, 11808–11812 (2002).
Tominaga, N. et al. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int. Immunol. 15, 1–10 (2003).
Brass, A.L., Kehrli, E., Eisenbeis, C.F., Storb, U. & Singh, H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10, 2335–2347 (1996).
Besnard, A.G. et al. NLRP3 inflammasome is required in murine asthma in the absence of aluminum adjuvant. Allergy 66, 1047–1057 (2011).
Eisenbarth, S.C., Colegio, O.R., O'Connor, W., Sutterwala, F.S. & Flavell, R.A. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453, 1122–1126 (2008).
De Nardo, D., De Nardo, C.M. & Latz, E. New insights into mechanisms controlling the NLRP3 inflammasome and its role in lung disease. Am. J. Pathol. 184, 42–54 (2014).
Kool, M. et al. An unexpected role for uric acid as an inducer of T helper 2 cell immunity to inhaled antigens and inflammatory mediator of allergic asthma. Immunity 34, 527–540 (2011).
Allen, I.C. et al. Analysis of NLRP3 in the development of allergic airway disease in mice. J. Immunol. 188, 2884–2893 (2012).
Kool, M. et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J. Immunol. 181, 3755–3759 (2008).
Gurung, P. et al. An NLRP3 inflammasome-triggered Th2-biased adaptive immune response promotes leishmaniasis. J. Clin. Invest. 125, 1329–1338 (2015).
Wang, H.W. & Joyce, J.A. Alternative activation of tumor-associated macrophages by IL-4: priming for protumoral functions. Cell Cycle 9, 4824–4835 (2010).
Elsasser-Beile, U. et al. Th1 and Th2 cytokine response patterns in leukocyte cultures of patients with urinary bladder, renal cell and prostate carcinomas. Tumour Biol. 19, 470–476 (1998).
Wise, G.J., Marella, V.K., Talluri, G. & Shirazian, D. Cytokine variations in patients with hormone treated prostate cancer. J. Urol. 164, 722–725 (2000).
Camp, B.J., Dyhrman, S.T., Memoli, V.A., Mott, L.A. & Barth, R.J. Jr. In situ cytokine production by breast cancer tumor-infiltrating lymphocytes. Ann. Surg. Oncol. 3, 176–184 (1996).
Zhang, W.J. et al. IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells. Cytokine 42, 39–47 (2008).
Roca, H. et al. IL-4 induces proliferation in prostate cancer PC3 cells under nutrient-depletion stress through the activation of the JNK-pathway and survivin up-regulation. J. Cell. Biochem. 113, 1569–1580 (2012).
Koller, F.L., Hwang, D.G., Dozier, E.A. & Fingleton, B. Epithelial interleukin-4 receptor expression promotes colon tumor growth. Carcinogenesis 31, 1010–1017 (2010).
Roth, F. et al. Aptamer-mediated blockade of IL4Ralpha triggers apoptosis of MDSCs and limits tumor progression. Cancer Res. 72, 1373–1383 (2012).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Bruchard, M. et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 19, 57–64 (2013).
Taylor, J., Schenck, I., Blankenberg, D. & Nekrutenko, A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics 2007, Chapter 10: Unit 10 15.
Ghiringhelli, F. et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat. Med. 15, 1170–1178 (2009).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Boeva, V., Lermine, A., Barette, C., Guillouf, C. & Barillot, E. Nebula–a web-server for advanced ChIP-seq data analysis. Bioinformatics 28, 2517–2519 (2012).
Iwasaki, T., Miyazaki, W., Rokutanda, N. & Koibuchi, N. Liquid chemiluminescent DNA pull-down assay to measure nuclear receptor-DNA binding in solution. Biotechniques 45, 445–448 (2008).
Acknowledgements
We thank V. Petrilli (INSERM 1052) for BALB/c Nlrp3−/− mice. Supported by the French National Research Agency (“Investissements d'Avenir” program; ANR-11-LABX-0021), the Ligue nationale contre le cancer (F.G. and F.V.), the Fondation de France (L.A.), the Institut National du Cancer (F.G.), the Fondation ARC (L.A.), the Conseil Régional de Bourgogne/INSERM (H.B.), the Centre National de la Recherche Scientifique, Fonds Européen de Développement Économique et Régional, Le Stadium, Orléans and Fondation pour la Recherche Médicale (F.G. and B.R.), the French National Research Agency (ANR-13-JSV3-0001 to L.A.), the Fondation pour l'aide à la Recherche sur la Sclérose en Plaques (L.A. and M.D.), the Ligue Régionale contre le cancer Comité Grand-Est (L.A.) and the European Commission (PCIG10-GA-2011-303719 to L.A.).
Author information
Authors and Affiliations
Contributions
M.B. did in vitro and in vivo experiments; C.R., V.D., A.H., F.C., H.B., A.C. and E.L. did in vitro experiments; D.T. and B.R. generated asthma experiments; R.B. performed and supervised the extraction, amplification and library preparation for RNA sequencing and ChIP-seq; E.H. did in vitro experiments; F.V. performed bioinformatics analysis; M.B., L.A., F.V. and F.G. supervised the study and wrote the manuscript; and F.G. conceived of and designed the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 NLRP3 is expressed in CD4+ T cells and is essential for TH2 differentiation.
(a)Western blot analysis of NLRP3 expression in T CD4+ cells from WT or Nlrp3−/– mice activated for 24 hours with anti-CD3 and anti-CD28 mAb. (b) Nlrp3 and Nlrp6 mRNA are expressed in T CD4+ T cells after TCR triggering. mRNA expression analysis of NLR genes in TH0, TH2, and TH1 differentiated from WT CD4+ T cells after 12h hours of stimulation with anti-CD3 and anti-CD28 mAb and the appropriate cytokines. (c) Ifng mRNA analysis in WT or Nlrp3−/– CD4+ T cells differentiated into TH1 or TH2 cells for 6 days. (d) Il4 mRNA analysis in WT or Nlrp3−/– CD4+ T cells differentiated into TH1 or TH2 cells for 6 days. (e) IL-13 production from WT or Nlrp3−/– CD4+ T cells differentiated into TH2 cells for 6 days were assessed by Elisa. (f) IL-5 production from WT or Nlrp3−/– CD4+ T cells differentiated in TH2 cells for 6 days were assessed by Elisa. (g) (Upper panel) Western blot analysis of GATA-3 expression in WT or Nlrp3−/– naïve and TH2 cells differentiated for 1, 3, or 6 days. (Lower panel) Quantification of GATA-3 expression. (h) WT or Nlrp3−/– CD4+ T cells were marked with CFSE and differentiated in TH2 cells for 3 days. Percentage of proliferating cells was assessed using CFSE dilution on day 1, 2 or 3. (i) Il4ra mRNA analysis in WT or Nlrp3−/– CD4+ T cells differentiated into TH0 or TH2 cells for 24h. (j) Ifng mRNA analysis in WT or Nlrp3−/– Balb/c CD4+ T cells differentiated into TH1 or TH2 cells for 3 days. (k) Il4m RNA analysis in WT or Nlrp3−/– Balb/c CD4+ T cells differentiated into TH1 or TH2 cells for 3 days.
Data are representative of three independent experiments *Indicates p<0.05
Supplementary Figure 2 Role of NLRP3 in TH1 and TH2 cells.
(a)IL1b mRNA analysis in Naïve T cells TH0 or TH2 cells after 3 days of differentiation. (b)Il4 mRNA analysis in TH0, TH2 and TH2 cultured with the Il-1 inhibitor anakinra or Nlrp3−/– after 3 days of differentiation. (c)Gata3 mRNA analysis in TH0, TH2 and TH2 cultured with the Il-1 inhibitor anakinra or Nlrp3−/– after 3 days of differentiation. (d) Caspase-1 activation was assessed by flow cytometry using FLICA-1 in naive T cells differentiated ex vivo in TH1 or TH2 either untreated or treated with Nigericin (Nig.10µM/ml) during the last hour. (e) IL-1β and IL-18 production was measured by ELISA in the same condition.
Data are representative of three independent experiments *Indicates p<0.05
Supplementary Figure 3 Subcellular localization of NLRP3.
(a)Frequency of cell with NLRP3 protein located in the nuclei or in the cytoplasm determined using immunofluorescence. (b) mRNA Expression of importin Ipo9, Ipo11 and Kpna2 in TH1 and TH2 cells after 3 days of differentiation
Data are representative of three independent experiments *Indicates p<0.05
Supplementary Figure 4 Global gene expression in wild-type or Nlrp3−/– T cells.
(a)Scatter plot displaying the RNA sequencing data of WT and Nlrp3−/– naive T cells. (b) Genes with similar expression in both Nlrp3−/– and WT Th2 were analyzed by using ClueGO software. Circles represent significantly enriched gene groups with p value corrected with Benjamini-Hochberg method.
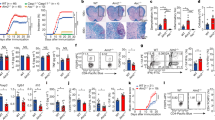
Supplementary Figure 5 NLRP3 interacts with IRF4 to dictate the TH2 program.
(a)Il4 promoter activity mesured in NIH-3T3 cells transfected with an il4 promoter driven luciferas reporter plasmid along with IRF4 and NLRP3 expression plasmids. (b) Western Blot analysis of IRF4 expression in TH0, TH1, TH2, Treg, TH17 after 24 hours differentiation. (c) Nlrp3 analysis by qPCR in EL4 T cell clone after 24 hours of treatment with PBS or with PMA/Ionomycin. (d) NLRP3 analysis by Western blotting in EL4 T cell clone after 24 hours of treatment with PBS or with PMA/Ionomycin. (e) Western blot analysis of GATA3 after an immunoprecipitation assay with anti-NLRP3 antibody in WT TH2 cells differentiated for two days. (f) Putative IRF4 binding sites on the il4 promoter found by Bioinformatic analysis. (g) ChIP assay of IRF4 binding on the IRF4 putative binding site 2 and 3 on the Il4 promoter in WT or Nlrp3−/–CD4+ T cells differentiated in TH0 or TH2 cells for one day. (h) ChIP assay of NLRP3 binding on the IRF4 putative binding site 2 and 3 on the Il4 promoter in WT or Nlrp3−/– CD4+ T cells differentiated in TH0 or TH2 cells for one day.
Data are representative of two independent experiments. *Indicates p<0.05
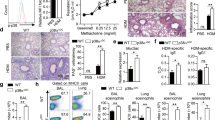
Supplementary Figure 6 NLRP3 deficiency impedes TH2 programs in an asthma model.
(a)OVA sensitized Rag−/– mice reconstituted with Nlrp3−/– or WT CD4 T cells were challenged three times with OVA in absence of aluminium adjuvant. Eosinophil (left panel) and lymphocyte (right panel) counts were determined in bronchoalveolar lavage fluid (BALF) 24h after the last intranasal OVA administration (mean +sd., 6 mice per group, 2 independent experiments).(b) Lung sections from Nlrp3−/–- or WT CD4 T cells were stained with periodic acid Schiff reagent (PAS) to visualize mucus. A semi-quantitative histological assessment of mucus hypersecretion (left panel) and inflammatory cell infiltration (right panel) was performed by two independent observers (mean +sd., 6 mice per group, 2 independent experiments).
Supplementary Figure 7 NLRP3 deficiency impedes TH2 programs in cancer models.
(a)mRNA analysis of Il4 and Il13 in mediastinal lymph nodes of WT or Nlrp3−/– mice naive or 12 days after intravenous injection of B16F10 melanoma cells. (b) mRNA analysis of Il4 and Il13 in mediastinal lymph nodes of WT or Asc−/– or Caspase-1−/– mice naive or 12 days after intravenous injection of B16F10 melanoma cells.
Data are representative of three independent experiments. *Indicates p<0.05
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 760 kb)
Supplementary Movie 1
NRLP3 localization in Th1 cells (AVI 64 kb)
Supplementary Movie 2
NRLP3 localization in Th2 cells (AVI 88 kb)
Rights and permissions
About this article
Cite this article
Bruchard, M., Rebé, C., Derangère, V. et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol 16, 859–870 (2015). https://doi.org/10.1038/ni.3202
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3202
This article is cited by
-
Tregs dysfunction aggravates postoperative cognitive impairment in aged mice
Journal of Neuroinflammation (2023)
-
Gut microbes exacerbate systemic inflammation and behavior disorders in neurologic disease CADASIL
Microbiome (2023)
-
Immunophenotype in acute exacerbation of chronic obstructive pulmonary disease: a cross-sectional study
Respiratory Research (2022)
-
Three important short-chain fatty acids (SCFAs) attenuate the inflammatory response induced by 5-FU and maintain the integrity of intestinal mucosal tight junction
BMC Immunology (2022)
-
Motoneuronal inflammasome activation triggers excessive neuroinflammation and impedes regeneration after sciatic nerve injury
Journal of Neuroinflammation (2022)