Abstract
Double-stranded DNA (dsDNA) in the cytoplasm triggers the production of interleukin 1β (IL-1β) as an antiviral host response, and deregulation of the pathways involved can promote inflammatory disease. Here we report a direct cytosolic interaction between the DNA-damage sensor Rad50 and the innate immune system adaptor CARD9. Transfection of dendritic cells with dsDNA or infection of dendritic cells with a DNA virus induced the formation of dsDNA-Rad50-CARD9 signaling complexes for activation of the transcription factor NF-κB and the generation of pro-IL-1β. Primary cells conditionally deficient in Rad50 or lacking CARD9 consequently exhibited defective DNA-induced production of IL-1β, and Card9−/− mice had impaired inflammatory responses after infection with a DNA virus in vivo. Our results define a cytosolic DNA-recognition pathway for inflammation and a physical and functional connection between a conserved DNA-damage sensor and the innate immune response to pathogens.
Similar content being viewed by others
Main
The appearance of double-stranded DNA (dsDNA) in the cytoplasm, which is normally a DNA-free environment, triggers potent inflammatory pathways that culminate in the production of interleukin 1β (IL-1β) and type 1 interferon. Such innate immune responses alert the host to the presence of danger and are important for the defense against viruses and bacteria1. To recognize the aberrant localization of DNA, the immune system uses several receptors for cytosolic DNA and associated signaling systems, which have in part overlapping and collaborative functions1. Central to the antiviral interferon response is the endoplasmic reticulum–associated adaptor STING, which is activated by the second messenger cGAMP, generated by the DNA sensor cGAS (cGAMP synthase)2,3. STING subsequently engages the kinase TBK1 to mediate phosphorylation and activation of the transcription factor IRF3 for the transcription of interferon-encoding genes4; in contrast, signaling by STING is largely dispensable for the generation of IL-1β5,6.
The highly proinflammatory cytokine IL-1β provides additional security checkpoints for antiviral immunity, particularly under circumstances in which viruses subvert the interferon system7,8. IL-1β also couples innate recognition of viruses to antiviral CD8+ T cell responses9,10, and as an endogenous pyrogen, IL-1β is responsible for fever reactions during infection. Because aberrant production of IL-1β can induce severely pathological conditions, its generation needs to be tightly controlled and involves at least two distinct signals11. The first signal for the generation of mature IL-1β triggers the transcriptional upregulation of pro-IL-1β mRNA, and the second signal results in the proteolytic processing of pro-IL-1β, typically by caspase-1, within inflammasomes. The specific inflammasome for DNA-induced processing of IL-1β contains the cytosolic DNA sensor AIM2 (ref. 1), which engages the common inflammasome adaptor ASC for the activation of caspase-1. The transcription of pro-IL-1β is mediated by the transcription factor NF-κB, which is retained in an inactive form in the cytosol in unstimulated cells via binding to inhibitory IκB proteins. The activation of NF-κB by most stimuli requires the IκB kinase (IKK)-mediated phosphorylation of IκB, which results in its ubiquitin-mediated degradation and the subsequent translocation of NF-κB dimers to the nucleus12. However, the exact mechanism by which the NF-κB signaling module is activated after cytosolic DNA is sensed is not well understood but is assumed to involve STING.
CARD9 is an adaptor specific to cells of the innate immune system that relays signals from pattern-recognition receptors to inflammatory responses13; it has an amino-terminal caspase-recruitment domain (CARD) for the recruitment of downstream effectors, as well as a coiled-coil region for protein oligomerization. Similar to most signaling adaptors14, CARD9 is multifunctionally engaged by various receptor systems depending on the specific context. Pattern-recognition receptors that signal via CARD9 include the transmembrane Syk-coupled C-type lectin receptors and such cytosolic sensors as RIG-I and Nod2 (refs. 15,16,17,18,19).
Here we report a direct cytosolic interaction between CARD9 and the DNA-binding protein Rad50, which is also prominently involved in the eukaryotic DNA-damage response20. Together with Mre11 and Nbs1, Rad50 can form a DNA receptor complex (the MRN complex) that detects double-strand breaks in DNA in the nucleus and subsequently triggers pathways needed to maintain genome integrity20. We found that cytoplasmic delivery of dsDNA by transfection of DNA or infection with a virus resulted in the formation of distinct dsDNA-Rad50-CARD9 complexes that selectively induced NF-κB signaling for IL-1β production. Our results define a DNA-recognition pathway for inflammation and demonstrate a previously unrecognized direct connection between an evolutionarily conserved sensor of DNA damage that translocates to the cytoplasm and an innate immune signaling system.
Results
Direct interaction of CARD9 with Rad50
To obtain insight into the role of CARD9 in innate immunity, we did a yeast two-hybrid screen with full-length CARD9 as the bait and searched for interaction partners in a cDNA library from human peripheral blood. The screening of 9.9 × 106 transformants yielded 33 bait-dependent interactors, which we analyzed further. The protein with the most frequent interactions with CARD9 in the yeast two-hybrid screen was Rad50 (data not shown).
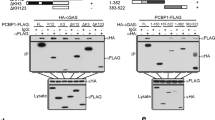
To confirm the potential direct CARD9-Rad50 association in a mammalian system, we did bioluminescence resonance energy transfer (BRET) experiments with COS-7 monkey kidney cells. As the transfer of energy from donor to acceptor in BRET analyses can occur only within a distance of less than 100 Å, protein-protein interactions can be analyzed in the environment of a living cell21. We subcloned sequence encoding fragments of CARD9 and Rad50 in frame with sequence encoding Renilla reniformis luciferase (Rluc) as an energy donor or yellow fluorescent protein (YFP) as an energy acceptor fluorophore21. In parallel, we generated Rluc and YFP fusion proteins with Bcl-10 (a protein known to interact with CARD9) as a positive control19, and also with the inflammasome DNA sensor AIM2 (ref. 1). We observed BRET ratios above the method-specific threshold for a binary protein-protein interaction21 for CARD9–Bcl-10 and CARD9-Rad50 but not for CARD9-AIM2 (Fig. 1a). Subsequent saturation experiments revealed a hyperbolic increase in BRET ratios, with increasing acceptor-to-donor ratios for CARD9 and Rad50 (Fig. 1b), which excluded the possibility of random bystander BRET. These biophysical data confirmed the association between CARD9 and Rad50 in mammalian cells (Fig. 1a,b). To identify the region in Rad50 that interacts with CARD9, we created several deletion mutants of Rad50 and did mapping experiments with BRET. The minimal region of Rad50 required for binding CARD9 was amino acids 628–786 (Fig. 1a), which is the structural domain that includes the zinc hook20.
(a) BRET assay of binary interactions of CARD9 with Rad50 fragments (A–D; identified at left), Bcl-10 or AIM2 as Rluc and YFP fusion proteins in COS-7 cells; dashed horizontal line, method-specific threshold for a positive protein-protein interaction. (b) BRET saturation experiments with cotransfected fusions of CARD9 and the Rad50 zinc hook (fragment B in a) or AIM2 at various acceptor/donor ratios (horizontal axis). (c,d) Immunoblot analysis (IB) of Rad50 and CARD9 in lysates of THP-1 cells without immunoprecipitation (Lysate) or after immunoprecipitation (IP) with antibody to CARD9 (anti-CARD9) (c) or anti-Rad50 (d) or the respective isotype-matched control antibody (Control), assessed with one gel and a single membrane that was cut for analysis. Right margin, molecular size in kilodalons (kDa). (e) Immunoblot analysis of Rad50 and CARD9 in lysates of THP-1 cells left untreated (−) or subjected to immunodepletion with anti-Rad50 one, two or three times (1–3 above lanes) (Lysates; left), and in lysates left untreated (−) or treated with anti-Rad50 three times (+), followed by incubation with beads coated with immobilized dsDNA (DNA beads) or streptavidin beads alone (Beads only) (right), assessed with one gel and a single membrane as in c,d. Data are from two independent experiments (a,b; mean and s.e.m.) or are representative of at least three independent experiments (c–e).
Next we investigated whether endogenous untagged CARD9 and Rad50 could bind to each other. Immunoprecipitation of proteins from THP-1 human monocytic cells showed that endogenous Rad50 immunoprecipitated together with endogenous CARD9 and vice versa (Fig. 1c,d). Because Rad50 has DNA-binding activity, we assessed the possibility that Rad50 might link recognition of dsDNA to binding to CARD9. Repeated treatment of lysates of THP-1 cells with antibody to Rad50 (three times) resulted in efficient depletion of Rad50 but did not affect the amount of CARD9 in these lysates (Fig. 1e). To precipitate DNA-associated proteins, we immobilized dsDNA containing genomic sequence from vaccinia virus (VV) onto agarose beads. Subsequent precipitation in the presence of Rad50 revealed that CARD9 specifically purified together with dsDNA-containing beads but not with empty control beads (Fig. 1e). CARD9 did not interact with dsDNA in the absence of Rad50 (Fig. 1e), which indicated that Rad50 was required for the binding of dsDNA to CARD9. Together these results indicated that Rad50 was able to bridge binding of DNA to the engagement of CARD9.
Rad50-CARD9 complexes sense dsDNA in the cytosol
We used confocal microscopy to visualize the associations among dsDNA, Rad50 and CARD9 in primary cells of the immune system and to investigate the cellular compartments in which these interactions occurred (Fig. 2a,b). Fluorescence immunostaining of endogenous proteins revealed that, as expected, Rad50 localized mainly to the nucleus in unstimulated bone marrow–derived dendritic cells (BMDCs)22, whereas CARD9 exhibited a cytoplasmic distribution pattern23 (Fig. 2a). The intracytoplasmic delivery of dsDNA resulted in the recruitment of Rad50 to dsDNA and in the formation of distinct dsDNA-Rad50 foci in the cytosol (Fig. 2a,b); these foci also contained Mre11 and Nbs1 (Fig. 2c and Supplementary Fig. 1a,b), which indicated sensing of cytoplasmic dsDNA by the entire MRN complex. CARD9 was also recruited to the dsDNA-MRN complex aggregates and specifically localized together with Rad50 (Fig. 2a,b). Quantitative analysis showed Rad50-CARD9 complexes in all cells that contained cytoplasmic DNA after transfection (Supplementary Fig. 1c,d). Cytoplasmic dsDNA-Rad50 complexes also formed in BMDCs from CARD9-deficient mice15 (data not shown and Supplementary Fig. 1e), which suggested that the detection of cytoplasmic dsDNA by Rad50 was independent of CARD9. These findings indicated that CARD9 was secondarily recruited to DNA-sensing Rad50 complexes and suggested that the Rad50-mediated engagement of CARD9 might represent a signal for DNA-mediated immune responses.
(a) Confocal microscopy of BMDCs left unstimulated (US) or transfected for 2 h with fluorescence-labeled (A647N) poly(dG:dC) (DNA) (2.5 μg/ml) and then stained with anti-Rad50 and anti-CARD9 and counterstained with the DNA-binding dye DAPI. Arrow (far right, bottom) indicates a cytosolic dsDNA-Rad50-CARD9 complex. (b) Confocal microscopy of BMDCs transfected with dsDNA, stained and analyzed as in a; the dsDNA-Rad50-CARD9 complex in outlined area at top left is presented at higher magnification (5.5×) and in various z-layers (z1–z5) in remaining images. (c) Confocal microscopy of BMDCs transfected with dsDNA as in a and stained with anti-Rad50, anti-Mre11 or anti-Nbs1 and counterstained with DAPI. Arrows indicate cytosolic dsDNA-Rad50-Mre11 (top) or dsDNA-Rad50-Nbs1 (bottom) complexes. Scale bars, 5 μm. Data are representative of at least three independent experiments with at least 50 cells per experiment and assay point.
CARD9 and Rad50 control DNA-induced IL-1β generation
To assess the potential functions of CARD9 complexes in DNA-induced innate immunity, we transfected BMDCs with various forms of DNA and measured cytokine production. The stimulation of wild-type BMDCs with linear synthetic poly(dA:dT), poly(dG:dC), purified genomic calf thymus DNA or circular bacterial plasmid DNA resulted in the robust production of mature IL-1β and type 1 interferon (Fig. 3a,b). In contrast, BMDCs that lacked CARD9 had considerable defects in IL-1β production after transfection of any of those DNA forms, although the production of IL-1β induced by the stimulation of Toll-like receptor 4 (TLR4) with lipopolysaccharide (LPS) or by the stimulation of TLR9 with CpG DNA was not affected (Fig. 3a); thus, CARD9-deficient cells did not have nonspecific impairment in the DNA-induced production of IL-1β. Moreover, the interferon responses controlled by the cGAS-STING pathway were also unaffected by the deletion of CARD9 (Fig. 3b), whereas STING-deficient BMDCs produced substantial amounts of IL-1β but no type I interferon (Fig. 3c,d), consistent with published data5,6. We therefore concluded that CARD9 was specifically required for DNA-induced IL-1β responses. This conclusion was supported by additional concentration-response and kinetic studies of CARD9-deficient cells stimulated with dsDNA (Fig. 3e,f).
(a,b) IL-1β (a) and IFN-β (b) in supernatants of wild-type (WT) and Card9−/− BMDCs left untreated (−) or transfected with dsDNA (1–4 μg/ml; wedges), in the form of poly(dA:dT) (dA:dT), poly(dG:dC) (dG:dC), purified genomic calf thymus DNA (CT-DNA) or circular bacterial plasmid DNA (Plasmid), or stimulated for 6 h with LPS, CpG or curdlan plus ATP. (c,d) IL-1β (c) and IFN-β (d) in supernatants of wild-type and STING-deficient (Tmem173−/−) BMDCs left untreated or transfected with dsDNA or stimulated with CpG plus ATP as in a,b. (e) IL-1β in supernatants of wild-type and Card9−/− BMDCs transfected for 0–24 h (horizontal axis) with poly(dG:dC) (4 μg/ml). (f) IL-1β in supernatants of wild-type and Card9−/− BMDCs left untreated or transfected for 16 h with increasing amounts (horizontal axis) of purified dsDNA from VV (VV-DNA). (g) IL-1β in supernatants of wild-type and Card9−/− BMDCs left untreated or transfected for 16 h with DNA from cowpox virus, VV or Escherichia coli. (h) Immunoblot analysis of DCs differentiated from Rad50Δ/ind bone marrow with (+) or without (−) the addition of 4-hydroxy-tamoxifen (4-OHT) to the culture on day 5, followed by lysis 4 or 5 d later (above lanes). β-actin serves as a loading control. (i) IL-1β in supernatants of BMDCs differentiated fromRad50+/ind or Rad50Δ/ind bone marrow as in h, followed by no further treatment or transfection with dsDNA (as in a) or stimulation with LPS or curdlan plus ATP 5 d after the addition of tamoxifen. ND, not detectable. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student's t-test). Data are from one experiment representative of at least three independent experiments (mean and s.e.m. in a–g,i).
To assess the biological relevance of those results, we transfected CARD9-deficient (Card9−/−) cells with purified microbial DNA from cowpox virus, VV or Escherichia coli. IL-1β responses to microbial genomic DNA were also specifically defective in Card9−/− BMDCs, although the interferon responses were intact (Fig. 3f,g and data not shown), indicative of a general role for signaling via CARD9 during cytosolic DNA–induced generation of IL-1β.
Next we investigated if Rad50 was required for the DNA-induced generation of IL-1β. Rad50 is essential for cell viability during division and embryonic or hematopoietic development, and deletion of Rad50 is embryonically lethal20,24,25. Therefore, we generated Rad50Δ/ind mice, which carry one Rad50-null allele (Rad50Δ) together with an inducibly deleted Rad50 allele (Rad50ind) and thus undergo conditional deletion of Rad50, as well as Rad50+/ind mice24, which carry one functional Rad50 allele and one Rad50ind allele. Each group of mice additionally ubiquitously expressed tamoxifen-inducible Cre recombinase (Rosa26-Cre-ERT2). We obtained bone marrow from those mice, differentiated the cells in culture and treated the differentiated, nondividing BMDCs with tamoxifen to activate Cre-mediated deletion of Rad50 in vitro. This protocol resulted in substantial depletion of Rad50 from BMDCs, although the protein was not completely absent, presumably due to its long half-life (Fig. 3h). In BMDCs that had undergone depletion of Rad50, stimulation with poly(dA:dT), poly(dG:dC) or mammalian genomic DNA resulted in IL-1β responses up to 80% lower than those of control (Rad50+/ind) cells that still had one functional Rad50 allele after Cre-mediated depletion of the Rad50ind allele (Fig. 3i). Nonetheless, in cells depleted of Rad50, parallel stimulation with CARD9-independent TLR agonists or the CARD9-dependent dectin-1 ligand curdlan induced strong IL-1β responses (Fig. 3i); thus, the cells depleted of Rad50 did not exhibit a general defect in the IL-1β response. Because Rad50 activates the kinase ATM after nuclear DNA damage is sensed20, we also assessed the involvement of ATM in the cytosolic DNA–induced generation of IL-1β. However, the production of IL-1β was not impaired by ATM deficiency26 (Supplementary Fig. 2). Together these genetic results established Rad50 as an innate immunological sensor of DNA and revealed that the Rad50- and CARD9-mediated DNA response operated independently of ATM to induce IL-1β.
Rad50 and CARD9 recruit Bcl-10 for NF-κB activation
We next investigated the mechanisms by which the Rad50-CARD9 interaction might mediate the generation of IL-1β. Because CARD9 is an activator of canonical NF-κB signaling13, we evaluated translocation of the NF-κB subunits RelA (p65) and c-Rel to the nucleus in Card9−/− cells into which we transfected DNA. By confocal microscopy, we observed that the activation of both NF-κB subunits was defective in Card9−/− BMDCs specifically after cytosolic transfection of DNA but not after stimulation of TLR9 mediated by CpG DNA (Fig. 4a–c). Consistent with those findings and the role of NF-κB activation in transcription of the gene encoding pro-IL-1β, the induction of pro-IL-1β mRNA and subsequent production of the pro-IL-1β polypeptide were also almost completely abrogated in Card9−/− BMDCs into which we had transfected DNA (Fig. 4d and Supplementary Fig. 3). Moreover, cytosolic DNA–induced transcription of genes encoding IL-6 and tumor-necrosis factor (TNF) was also much lower in Card9−/− cells than in wild-type cells (Supplementary Fig. 4a,b). However, in agreement with the unaffected interferon responses noted above (Fig. 3), the activation of IRF3 was intact in Card9−/− cells, as assessed by both phosphorylation of IRF3 and translocation of IRF3 to the nucleus (Fig. 4e,f). In contrast, in STING-deficient cells, the activation of IRF3 was abolished (Fig. 4g) and transcription of the gene encoding interferon-β (IFN-β) was completely blocked (data not shown). When we transfected Card9−/− or STING-deficient BMDCs with cGAMP, which is a strong inducer of type 1 interferon that directly activates STING2,3, we also found that STING was essential for the interferon response2,3, while CARD9 was dispensable (Supplementary Fig. 5a,b). Next, we investigated the role of STING in DNA-induced IL-1β generation in more detail. Consistent with the almost intact IL-1β production in STING-deficient cells stimulated with DNA5,6 (Fig. 3c), NF-κB-dependent transcription of the gene encoding pro-IL-1β was not diminished in STING-deficient cells, and by confocal microscopy we also detected substantial translocation of both p65 and c-Rel to the nucleus in STING-deficient BMDCs (Fig. 4h,i). Together these data indicated that CARD9 was essential for a specific pathway that mediated the activation of NF-κB for the generation of pro-IL-1β in response to sensing of cytosolic DNA. Moreover, although STING contributed to optimal NF-κB responses, it was largely dispensable for the induction of transcription of the gene encoding pro-IL-1β after detection of DNA.
(a) Confocal microscopy of wild-type and Card9−/− BMDCs left untreated (UT), transfected with poly(dG:dC) (DNA) (2.5 μg/ml) or stimulated with CpG for 1 h, then fixed and stained with anti-p65 or anti-c-Rel and counterstained with DAPI. Scale bar, 5 μm. (b,c) Quantification of the translocation of p65 (b) and c-Rel (c) into the nucleus of BMDCs (n ≥ 100) treated and visualized as in a. (d) Quantitative real-time PCR analysis of IL-1β transcripts in wild-type and Card9−/− BMDCs left untreated, transfected with poly(dG:dC) (DNA) (2.5 μg/ml) or stimulated with CpG for 4 h; results are presented relative to those of β-actin mRNA. *P < 0.001 (Student's t-test). (e) Immunoblot analysis of IRF3 phosphorylated at Ser396 (p-IRF3) and total IRF3 in cytosolic extracts of wild-type and Card9−/− BMDCs left untransfected (−) or transfected for 10–120 min (above lanes) with dsDNA. (f) Immunoblot analysis of IRF3 and lamin B (loading control) in nuclear extracts of BMDCs treated as in e. (g) Immunoblot analysis of wild-type and Tmem173−/− BMDCs stimulated and analyzed as in e. (h) Translocation of p65 (left) and c-Rel (right) into the nucleus of wild-type or Tmem173−/− BMDCs left untreated or transfected with dsDNA, analyzed as in b,c. (i) Quantitative real-time PCR analysis of IL-1β transcripts in wild-type and Tmem173−/− BMDCs treated and analyzed as in d. Data are representative of three (a–h) or two (i) independent experiments (mean and s.e.m. in d,i).
CARD9 engages Bcl-10 to activate NF-κB after the ligation of C-type lectin receptors or RIG-I15,16,18,19. Thus, we stimulated Bcl10−/− BMDCs with poly(dA:dT), poly(dG:dC), purified genomic calf thymus DNA or circular plasmid DNA to study the involvement of Bcl-10 during the DNA-induced innate immune response. Bcl-10 was essential for the activation of NF-κB and generation of IL-1β induced by cytosolic dsDNA but not for the production of IL-1β induced by the stimulation of TLR4 or TLR9 (Fig. 5a). Moreover, similar to CARD9, Bcl-10 was essential for the upregulation of transcription of the gene encoding pro-IL-1β but was dispensable for signaling via IRF3 and synthesis of IFN-β (Fig. 5b,c, Supplementary Fig. 3 and data not shown). To investigate the roles of CARD9 and Bcl-10 in inflammasome activation, we also assessed caspase-1 activation in CARD9- or Bcl-10-deficient BMDCs. This response, which is mediated by AIM2 and ASC1, was intact in the absence of CARD9 or Bcl-10 (Fig. 5d), which indicated that CARD9 and Bcl-10 specifically controlled the signal for the generation of pro-IL-1β.
(a,b) IL-1β (a) and IFN-β (b) in supernatants of wild-type and Bcl10−/− BMDCs left untreated or transfected with dsDNA (1–4 μg/ml) of various origins (as in Fig. 2a) or stimulated with LPS, CpG or curdlan plus ATP (control). (c) Quantitative real-time PCR analysis of IL-1β transcripts in BMDCs left untreated, transfected with poly(dG:dC) (DNA) (2.5 μg/ml) or stimulated with CpG for 4 h (presented as in Fig. 4d). (d) Immunoblot analysis of mature caspase-1 (p10) in culture supernatants of BMDCs of various genotypes (above lanes) left untreated, transfected with poly(dG:dC) (DNA) or stimulated with LPS plus ATP for 8 h. (e) Confocal microscopy of wild-type, Card9−/− and Bcl10−/− BMDCs (n ≥ 50 cells per experiment and assay point) with at least 50 cells per experiment and assay point transfected with poly(dG:dC) (DNA) for 2 h and then stained with anti-Rad50 and anti-CARD9 or anti-Bcl-10 and counterstained with DAPI (merged images); arrows indicate cytosolic dsDNA-Rad50-CARD9, dsDNA–Rad50–Bcl-10, or dsDNA-Rad50 complexes. Scale bar, 5 μm. *P < 0.01 and **P < 0.001 (Student's t-test). Data are representative of at least three independent experiments (mean and s.e.m. in a–c).
On the basis of the findings that CARD9 and Bcl-10 acted together to activate NF-κB after cytosolic DNA was sensed, we next investigated whether Bcl-10 was recruited to dsDNA-Rad50 complexes. By confocal microscopy we observed that endogenous Bcl-10 localized together with dsDNA-Rad50 aggregates after transfection of DNA (Fig. 5e). Moreover, that recruitment of Bcl-10 to cytosolic Rad50 was mediated via CARD9, as we did not observe it in CARD9-deficient cells. Additionally, CARD9 was recruited to the dsDNA-Rad50 complexes in Bcl10−/− cells as it was in wild-type cells, which demonstrated that the initial steps of formation of the Rad50-CARD9 complex did not require the effector Bcl-10. Thus, the sensing of cytosolic DNA by Rad50 and the subsequent assembly of Rad50-CARD9 complexes resulted in further recruitment of Bcl-10 for the activation of NF-κB.
Rad50-CARD9 senses cytosolic viral DNA upon infection
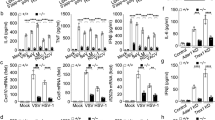
Having established the critical functions of Rad50, CARD9 and Bcl-10 in the DNA-induced production of IL-1β, we investigated the roles of these factors during viral infection. We used the poxvirus vaccinia virus (VV) as a model because the life cycle of poxviruses includes DNA replication in the cytoplasm27. Similar to the results reported above obtained by transfection of DNA, infection of wild-type or Card9−/− BMDCs with VV resulted in the localization of Rad50 to the cytoplasmic viral DNA (Fig. 6a,b and data not shown), which confirmed that this DNA sensor was able to detect cytosolic viral infection. In wild-type BMDCs, CARD9 was also recruited to the VV-dsDNA-Rad50 foci, which resulted in the formation of VV-dsDNA-Rad50-CARD9 complexes (Fig. 6a,b) that were morphologically similar to those observed after transfection of synthetic dsDNA (Fig. 2). To investigate whether sensing a DNA virus also induced signaling via CARD9, we measured inflammatory cytokines after infecting wild-type, Card9−/− and Bcl10−/− BMDCs with VV and found that both Card9−/− BMDCs and Bcl10−/− BMDCs demonstrated severely impaired IL-1β production upon infection with VV in vitro (Fig. 6c), which suggested that recognition of VV activated signaling via CARD9. Similarly, the VV-induced generation of the NF-κB-controlled cytokines TNF and IL-6 was also considerably impaired in Card9−/− BMDCs and Bcl10−/− BMDCs (Fig. 6c). Control infection of cells with the RNA virus vesicular stomatitis virus resulted in defective IL-1β production in Card9−/− cells (Supplementary Fig. 6), in accord with the function of CARD9 downstream of RIG-I17,18,28,29.
(a,b) Confocal microscopy of wild-type BMDCs infected with VV, then fixed and stained with anti-Rad50 and anti-CARD9 and counterstained with DAPI. Arrows (a) indicate viral dsDNA–Rad50–CARD9 complexes; viral dsDNA–Rad50–CARD9 complex in outlined area at top left (b) is presented at higher magnification (5.5×) and in various z-layers in other images. Scale bars, 5 μm. (c) IL-1β, TNF and IL-6 in supernatants of wild-type, Card9−/− and Bcl10−/− BMDCs left uninfected (UI) or infected with VV at a multiplicity of infection (MOI) of 0.1, 1 or 10. *P < 0.05, **P < 0.01 and ***P < 0.001 (Student's t-test). Data are from one experiment representative of three independent experiments (a,b) or are representative of three experiments (c; mean and s.e.m.).
To investigate the physiological importance of CARD9 in innate immune responses to infection with a DNA virus in vivo, we infected Card9−/− mice with VV via an intravenous route. After 6 h, we measured the serum concentrations of IL-1β by cytometric bead array, which revealed that the Card9−/− mice exhibited defective IL-1β production upon infection (Fig. 7a). Because IL-1β regulates adaptive antiviral CD8+ T cell responses9,10, we investigated the frequency of IFN-γ-producing viral antigen–specific CD8+ T cells 8 d after infecting wild-type and Card9−/− mice with VV. Consistent with the diminished IL-1β production observed in Card9−/− mice, antiviral CD8+ T cell responses were also significantly impaired in the absence of CARD9 (Fig. 7b). Thus, the Rad50-CARD9 complexes sensed viral cytoplasmic DNA after cell infection, and activation of signaling via CARD9 was critical for the subsequent host response in vivo.
(a) IL-1β in the serum of wild-type and Card9−/− mice 6 h after infection with VV in vivo. (b) Frequency of IFN-γ+CD8+ T cells among splenocytes isolated from wild-type and Card9−/− mice 8 d after infection with VV in vivo and stimulated for 5 h with a VV-specific peptide (B8R) or a control peptide (ovalbumin amino acids 257–264 (OVA(257–264)), assessed by flow cytometry. Each symbol represents an individual mouse; small horizontal lines indicate the mean (±s.e.m.). *P < 0.05 and **P < 0.01 (Student's t-test). Data are representative of one experiment.
Discussion
Understanding the recognition of cytosolic DNA and downstream signaling has been a focus of intense research for several years. Many sensors of cytosolic DNA have been described, including AIM2, cGAS, DAI, LRRFIP1, IFI16, DHX9, DDX36, DDX41 and proteins with known functions in the DNA-damage response1,2,4. Although the physiological roles of some of these receptors need to be defined genetically, experiments with gene-deficient mice have revealed that the cGAS-STING signaling cascade is the key regulator of the IRF3-mediated interferon response4,5,30. Nonetheless, disruption of STING did not affect the generation of IL-1β after infection with a DNA virus, although it almost completely abolished the DNA-induced production of INF-β5, and we also observed that STING was largely dispensable for transcription of the gene encoding pro-IL-1β and the generation of IL-1β.
Data obtained with CARD9- or Bcl-10-deficient BMDCs revealed that CARD9–Bcl-10 complexes specifically controlled NF-κB activation and transcription of the gene encoding pro-IL-1β after cytosolic DNA was sensed. Transcription of the genes encoding the NF-κB-dependent cytokines TNF and IL-6 was also significantly lower in Card9−/− cells stimulated with DNA, and Card9−/− DCs showed impaired production of TNF and IL-6 upon infection with a DNA virus. However, because the generation of TNF and IL-6 was not completely defective in Card9−/− cells, it is possible that STING-dependent signaling might contribute to the production of certain NF-κB-dependent factors other than pro-IL-1β. It remains to be determined whether and how distinct NF-κB responses could be regulated differently via CARD9 and, potentially, STING. Because Card9−/− cells and Bcl10−/− cells exhibited normal IRF3 activation and interferon production, it is unlikely that CARD9–Bcl-10 complexes would directly influence the cGAS-STING cascade. This hypothesis is supported by the finding that triggering STING with cGAMP induced normal production of IFN-β in Card9−/− DCs. Thus, the Rad50-CARD9 signaling pathway regulates specifically NF-κB-dependent innate immune responses required for transcription of the gene encoding pro-IL-1β. The essential function of AIM2 in DNA-mediated cleavage of pro-IL-1β1 has been confirmed31,32. Our findings and those data together indicate a comprehensive mechanism for the DNA-induced generation of IL-1β in which CARD9 complexes can mediate the generation of pro-IL-1β, which is subsequently processed by the AIM2 inflammasome. Because CARD9 is expressed selectively in myeloid cells13 and because deletion of CARD9 did not completely abolish IL-1β production, CARD9-independent mechanisms can apparently contribute to the generation of IL-1β, potentially as a failsafe mechanism.
Rad50 senses DNA via its amino- and carboxy-terminal nucleotide-binding domains, which associate with Mre11 and Nbs1 to form a globular DNA-binding complex20. Our deletion of Rad50 mediated by Cre and loxP has provided the first genetic confirmation, to our knowledge, of an essential function for Rad50 in signaling by the innate immune system. CARD9 associated directly with Rad50. That association involved the Rad50 zinc-hook region, which is separated from the Rad50 DNA-binding domain by a coiled-coil domain approximately 50 nm in length. Bcl-10 was also recruited to dsDNA-Rad50 complexes via CARD9. We therefore propose a model in which, together with Mre11 and NBS1, Rad50 detects viral or transfected dsDNA in the cytosol, which results in the recruitment of CARD9 to the Rad50 zinc-hook region and the subsequent engagement of Bcl-10 for inflammatory downstream signal transduction. Identification of the mechanism through which Rad50 localizes to the cytosol will require further investigation. We speculate that these mechanisms could involve modifications of Nbs1 because this factor controls the subcellular localization of the MRN complex, as demonstrated by the finding that the nuclear concentrations of Mre11 and Rad50 are lower in fibroblasts that contain truncation mutants of Nbs1 than in cells that contain wild-type Nbs1 and that the concentrations of Mre11 and Rad50 are higher in the cytoplasm of the fibroblasts with truncated Nbs1 (ref. 33).
The MRN complex has been reported to localize to sites of viral replication and to elicit antiviral defense mechanisms involving the concatemerization of viral DNA and, potentially, other pathways, including the activation of STING34,35,36, although it is still unclear how Rad50 would couple to the cGAS-STING cascade. Additional DNA-damage–control factors, such as DNA-PK and Ku70 (refs. 37,38), have also been linked to the sensing of cytosolic DNA for innate immunity. Together these studies demonstrate a more global interaction between the evolutionarily conserved DNA-damage response system and the innate immune response to pathogens. By demonstrating a direct physical and functional connection between the damage sensor Rad50 and the proinflammatory signaling adaptor CARD9, we have now provided the first mechanistic insight into these interactions, to our knowledge.
IL-1β production induced by cytosolic DNA has a critical role in host defense31,32,39. Therefore, IL-1β provides an antiviral selective pressure, a proposal highlighted by the observation that various viruses have evolved strategies to inhibit the production of IL-1β by interfering with the NF-κB signaling pathway at multiple steps8 or by inhibiting IL-1β signaling, for example, through the expression of soluble IL-1β receptors that prevent the fever reaction7,8,10,40. Several DNA viruses, such as adenovirus, have also developed strategies to inhibit Rad50 signaling, which suggests that the Rad50 pathway is potentially subverted by various viruses34,41. As indicated above, IL-1β not only is important for immunological defense but also is an important factor in autoinflammation11. Endogenous DNA that is inappropriately cleared can accumulate in cytosolic compartments and drive inflammatory diseases associated with increased concentrations of interferon and IL-1β42,43,44. Such findings indicate that it will be important to investigate the contributions of signaling via Rad50-CARD9 to autoinflammatory conditions associated with responses to cytosolic DNA. Ultimately, future studies of the Rad50-CARD9 pathway may lead to selective strategies for dampening DNA-induced IL-1β-driven inflammatory responses without compromising antiviral interferon production.
Methods
Mice.
Mice deficient in CARD9 (Card9−/−)15, Bcl-10 (Bcl10−/−)45, STING (Tmem173−/−)46, ATM (Amt−/−)26 or ASC (Pycard−/−)47, and Rad50Δ/ind × Rosa26-CreERT2 mice24 and Rad50+/ind × Rosa26-CreERT2 mice24 were used at 6–12 weeks of age. Animal experiments were approved by Regierung von Oberbayern.
Media and reagents.
All reagents, including poly(dG:dC) (poly(dG-dC) • poly(dG-dC) acid sodium salt; P9389), calf thymus DNA (D4764) and poly(dA:dT) (poly(dA-dT) • poly(dA-dT) acid sodium salt; P0883) were from Sigma, if not stated otherwise. cGAMP (tlrl-cga-s) was from Invivogen. The pmaxGFP vector from Lonza was used as the circular plasmid DNA. Cell culture reagents were from Invitrogen, and FCS was from HyClone. Mouse recombinant granulocyte-macrophage colony-stimulating factor was from PreproTech. CpG (oligonucleotide 1826), ultrapure LPS and endotoxin-free DNA from E. coli strain K12 were from InvivoGen, and curdlan was from Wako. Poxvirus genomic DNA from VV strain CVA and from cowpox virus (isolate 81/01, fifth passage in MA104 monkey kidney cells; originally provided by S. Essbauer) was isolated and purified from infected cell cultures as described48.
Cell culture and stimulation.
COS-7 cells, THP-1 cells and BMDCs were cultured as described18,21. For transfection of DNA, Lipofectamine 2000 was used according to the manufacturer's protocol (Invitrogen). Unless stated otherwise, LPS (100 ng/ml), CpG (1 μM), curdlan (200 μg/ml) and ATP (5 mM) were used for stimulation. ATP was added 45 min before the end of the experiment. Bone marrow from Rad50Δ/ind × Rosa26-CreERT2 mice and Rad50+/ind × Rosa26-CreERT2 mice was cultured for 5 d in the presence of granulocyte-macrophage colony-stimulating factor before the addition of 1 μM 4-hydroxy-tamoxifen and then were cultured for another 4–5 d before analysis.
Yeast two-hybrid screen.
The yeast two-hybrid screen with CARD9 as the bait was done by Dualsystems Biotech AG. For generation of the bait construct, cDNA encoding human CARD9 isoform 2 was subcloned into the vector pLexA-DIR, then the bait was transformed together with a human adult peripheral blood cDNA library into yeast host strain NMY32. A total of 9.9 × 106 transformants were screened, which yielded 96 transformants that grew on selective medium. The β-galactosidase activity of positive transformants was assessed by PXG β-galactosidase assay, and 77 (corresponding to 33 distinct genes) of the 96 initial positive transformants showed β-galactosidase activity and were considered to be truly positives. The library plasmids were isolated from positive clones, and the identity of positive interactors was determined by sequencing: 19 of the 77 clones contained Rad50 sequences.
BRET analysis.
BRET measurements were made in transfected COS-7 cells as described21. Full-length open reading frames of genes encoding CARD9 (National Center for Biotechnology Information reference sequence NM_052814), Bcl-10 (NM_003921) and AIM2 (NM_004833) and fragments of Rad50 (NM_005732) were introduced into plasmid vectors for the expression of amino- and carboxy-terminal fusions with Rluc or YFP. The truncated Rad50 constructs were generated by PCR. All eight possible combinations of Rluc (donor) or YFP (acceptor) fusions tagged at the amino or carboxyl terminus were tested for each putative interaction pair at an acceptor/donor ratios of 3:1, unless indicated otherwise. BRET ratios were calculated by the equation R = (IA / ID) − cf, where R is the BRET ratio, IA is the BRET signal, ID is the Rluc signal and cf is a correction factor ((IA / ID)control), with cotransfection of the donor fusion protein with YFP in the absence of the second protein of interest used as the control. The method-specific threshold for a positive protein-protein interaction was determined as a BRET ratio of 0.094 for the donor/acceptor combination that resulted in the highest BRET ratio, as described21.
Coimmunoprecipitation and immunoblot analysis.
THP-1 cells were lysed and proteins were immunoprecipitated as described18. Cell lysates or cell supernatants were subjected to standard immunoblot analysis techniques as described15. Proteins in cell-free supernatants were extracted by methanol-chloroform precipitation. Cytosolic and nuclear extracts were prepared as described45.
Antibodies.
The primary antibodies anti-CARD9 (sc-99054), anti-Rad50 (sc-56209 and sc-74460), anti-caspase-1 p10 (sc-514), anti-Lamin B (sc-6217), anti-NF-κB p65 (sc-372), and anti-cRel (sc-71) were from Santa Cruz, and anti-Bcl-10 (4237), anti-Mre11 (4895), anti-p95 (anti-Nbs1; 3002), anti-IRF-3 (4302) and antibody to IRF3 phosphorylated at Ser396 (4947) were from Cell Signaling. Anti-CARD9 (rabbit polyclonal antibody raised against an amino-terminal peptide of CARD9) was provided by M. Thome. Secondary donkey anti-mouse (A21203) and goat anti-rabbit (A11008) were conjugated to Alexa Fluor 594 and Alexa Fluor 488 (Molecular Probes), respectively, for immunofluorescence.
Nucleic acid affinity purification.
Oligonucleotides (50–base pair DNA) containing a partial sequence of the terminal repeats of VV genomic DNA were synthesized by Biomers (sense, 5′-CCATCAGAAAGAGGTTTAATATTTTTGTGAGACCATCGAAGAGAGAAAGA-3′, and antisense, 5′-TCTTTCTCTCTTCGATGGTCTCACAAAAATATTAAACCTCTTTCTGATGG-3′), then were annealed to generate dsDNA and were immobilized on Strep-Tactin Superflow resin (IBA). Nucleic acid affinity purification was done for lysates of THP-1 cells as described49. For the immunodepletion of Rad50, THP-1 lysates were incubated for up to three consecutive rounds with anti-Rad50 (sc-56209; Santa Cruz) or isotype-matched control antibody (Mouse IgG1 Isotype Control; MAB002; R&D Systems) immobilized on Sepharose beads.
Immunofluorescence staining and confocal microscopy.
Immunofluorescence staining was done by standard technology as described15. Images were obtained with a TCS SP5 AOBS confocal laser-scanning microscope (Leica) with a Plan-Apochromat 63× oil-immersion objective with a numerical aperture of 1.4. For presentation, images were processed with ImageJ software (US National Institutes of Health) with linear contrast enhancement on entire images.
DNA labeling by nick translation.
The direct labeling of double-stranded poly(dG:dC) DNA by incorporation of ATTO 647N fluorescence–labeled aminoallyl-dUTP nucleotides (Jena Bioscience) was achieved by nick translation as described50.
Cytokine measurement.
Cytokine concentrations were measured by enzyme-linked immunosorbent assay (BD Biosciences, eBioscience or PBL Biomedical Laboratories) or by cytometric bead array (CBA; BD Biosciences) according to the manufacturer's instructions.
Real-time PCR.
Total RNA was isolated and transcribed using standard methods. The specific primer pairs were as follows: IL-1β, 5′-TGTAATGAAAGACGGCACACC-3′ and 5′-TCTTCTTTGGGTATTGCTTGG-3′; TNF, 5′-TCTTCTCATTCCTGCTTGTGG-3′ and 5′-GGTCTGGGCCATAGAACTGA-3′; IL-6, 5′-GCTACCAAACTGGATATAATCAGGA-3′ and 5′-CCAGGTAGCTATGGTACTCCAGAA-3′; β-actin, 5′-AGACCTCTATGCCAACACAG-3′ and 5′-TCGTACTCCTGCTTGCTGAT-3′. The qPCR Core kit for SYBR Green I (Eurogentec) and a LightCycler 480 Real-Time PCR System were used as indicated by the manufacturer. IL-1β mRNA expression was calculated as the ratio of the real-time PCR signal of IL-1β mRNA to that of the β-actin mRNA and was normalized to a wild-type unstimulated control.
Viral infections.
VV strain CVA and vesicular stomatitis virus Indiana (Mudd-Summer strain) were used for viral infection. VV was propagated in CV-1 cells and was purified twice by ultracentrifugation through 36% sucrose cushions; viral titers were measured by plaque assay of CV-1 cells with crystal violet as described48. Vesicular stomatitis virus was propagated in BHK-21 baby hamster kidney cells as described18. In vitro infection was done at a multiplicity of infection of 0.1–10. For in vivo experiments, mice were given intravenous injection of 1 × 107 plaque-forming units of virus in 200 μl of PBS. After 6 h, the serum was collected to determine the IL-1β concentration. Researchers were 'blinded' to sample identity for injection and serum collection of mice. Splenocytes from vaccinated mice were isolated at 8 d after infection and were stimulated for 5 h at 37 °C with the H-2Kb-restricted VV-specific peptide B8R (amino acids 20–27 (TSYKFESV); 1 μg/ml) or the control peptide ovalbumin (amino acids 257–264 (SIINFEKL); 1 μg/ml) in the presence of 1 μg/ml brefeldin A (Sigma-Aldrich). The cells were stained as live or dead with ethidium monoazide bromide (Invitrogen) and nonspecific binding was blocked with Fc-Block (anti-CD16/CD32; BD Biosciences); the cell surface was stained with an eFluor450-conjugated anti-CD8α (53-6.7; eBioscience). For intracellular staining of IFN-γ, fluorescein isothiocyanate–conjugated anti-IFNγ (XMG1.2; BD Biosciences) was used with a Cytofix/Cytoperm kit according to the manufacturer's instructions (BD Biosciences). Data were acquired with a FACSCanto II (BD Biosciences) and were analyzed with FlowJo software (TreeStar).
Statistics.
P values were calculated with a two-tailed Student's t-test for independent samples with Microsoft Excel.
References
Paludan, S.R. & Bowie, A.G. Immune sensing of DNA. Immunity 38, 870–880 (2013).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Goubau, D., Deddouche, S. & Reis, E.S.C. Cytosolic sensing of viruses. Immunity 38, 855–869 (2013).
Ishikawa, H., Ma, Z. & Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Zhang, Z. et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 (2011).
Epperson, M.L., Lee, C.A. & Fremont, D.H. Subversion of cytokine networks by virally encoded decoy receptors. Immunol. Rev. 250, 199–215 (2012).
Smith, G.L. et al. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J. Gen. Virol. 94, 2367–2392 (2013).
Staib, C., Kisling, S., Erfle, V. & Sutter, G. Inactivation of the viral interleukin 1beta receptor improves CD8+ T-cell memory responses elicited upon immunization with modified vaccinia virus Ankara. J. Gen. Virol. 86, 1997–2006 (2005).
Zimmerling, S., Waibler, Z., Resch, T., Sutter, G. & Schwantes, A. Interleukin-1β receptor expressed by modified vaccinia virus Ankara interferes with interleukin-1β activity produced in various virus-infected antigen-presenting cells. Virol. J. 10, 34 (2013).
Dinarello, C.A., Simon, A. & van der Meer, J.W. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 11, 633–652 (2012).
Vallabhapurapu, S. & Karin, M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 (2009).
Roth, S. & Ruland, J. Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol. 34, 243–250 (2013).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Gross, O. et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 442, 651–656 (2006).
Hara, H. et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat. Immunol. 8, 619–629 (2007).
Hsu, Y.M. et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 8, 198–205 (2007).
Poeck, H. et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nat. Immunol. 11, 63–69 (2010).
Strasser, D. et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-δ to elicit Card9 adaptor-mediated innate immunity. Immunity 36, 32–42 (2012).
Stracker, T.H. & Petrini, J.H. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011).
Gersting, S.W., Lotz-Havla, A.S. & Muntau, A.C. Bioluminescence resonance energy transfer: an emerging tool for the detection of protein-protein interaction in living cells. Methods Mol. Biol. 815, 253–263 (2012).
Maser, R.S., Monsen, K.J., Nelms, B.E. & Petrini, J.H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17, 6087–6096 (1997).
Goodridge, H.S. et al. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J. Immunol. 182, 1146–1154 (2009).
Adelman, C.A., De, S. & Petrini, J.H. Rad50 is dispensable for the maintenance and viability of postmitotic tissues. Mol. Cell. Biol. 29, 483–492 (2009).
Luo, G. et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96, 7376–7381 (1999).
Barlow, C. et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86, 159–171 (1996).
Roberts, K.L. & Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 16, 472–479 (2008).
Abdullah, Z. et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31, 4153–4164 (2012).
Kok, K.H. et al. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9, 299–309 (2011).
Li, X.D. et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Fernandes-Alnemri, T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393 (2010).
Rathinam, V.A. et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 (2010).
Carney, J.P. et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–486 (1998).
Lilley, C.E., Schwartz, R.A. & Weitzman, M.D. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15, 119–126 (2007).
Weitzman, M.D., Carson, C.T., Schwartz, R.A. & Lilley, C.E. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst.) 3, 1165–1173 (2004).
Kondo, T. et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA 110, 2969–2974 (2013).
Ferguson, B.J., Mansur, D.S., Peters, N.E., Ren, H. & Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 1, e00047 (2012).
Zhang, X. et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 186, 4541–4545 (2011).
Saiga, H. et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int. Immunol. 24, 637–644 (2012).
Alcamí, A. & Smith, G.L. A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. USA 93, 11029–11034 (1996).
Stracker, T.H., Carson, C.T. & Weitzman, M.D. Adenovirus oncoproteins inactivate the Mre11-Rad50–NBS1 DNA repair complex. Nature 418, 348–352 (2002).
Boswell, J.M., Yui, M.A., Burt, D.W. & Kelley, V.E. Increased tumor necrosis factor and IL-1β gene expression in the kidneys of mice with lupus nephritis. J. Immunol. 141, 3050–3054 (1988).
Dombrowski, Y. et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3, 82ra38 (2011).
Popovic, K. et al. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis Rheum. 52, 3639–3645 (2005).
Ruland, J. et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closure. Cell 104, 33–42 (2001).
Jin, L. et al. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 187, 2595–2601 (2011).
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004).
Gasteiger, G., Kastenmuller, W., Ljapoci, R., Sutter, G. & Drexler, I. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J. Virol. 81, 11925–11936 (2007).
Bürckstümmer, T. et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266–272 (2009).
Cremer, M. et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 463, 205–239 (2008).
Acknowledgements
We thank M. Thome (University of Lausanne) for the antibody to CARD9; S. Essbauer (Bundeswehr Institute of Microbiology) for cowpox virus; and R. Ljapoci for technical assistance. Supported by the Bavarian Genome Research Network (A.C.M.), the Ludwig-Maximilians-Universität Excellence Initiative (42595-6 to A.C.M.), Deutsche Forschungsgemeinschaft (SFB684 to H.L. and J.R. and SFB1054 to J.R.), the US National Institutes of Health (U19AI083025 to K.-P.H. and GM56888 to J.H.J.P.) and the European Research Council (J.R.).
Author information
Authors and Affiliations
Contributions
S.R., A.R., H.L. and J.R. designed the study; S.R., A.R., A.S.L.-H., V.L., A.M. and K.V. did the experiments; S.R., A.R., A.S.L.-H., S.W.G., A.C.M., K.-P.H., I.D., H.L. and J.R. analyzed the results; S.R., A.R. and A.S.L.-H. generated the figures; L.J. and J.H.J.P. provided reagents; and S.R. and J.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 MRN and Rad50-CARD9 complex formation at cytosolic dsDNA.
(a, b) BMDCs were transfected with dsDNA for 2 hours, stained with DAPI and anti-Rad50 and anti-Mre11 or anti-Nbs1 antibodies, and analyzed using confocal microscopy. The dsDNA/Rad50/Mre11 or dsDNA/Rad50/Nbs1 complexes (boxes) were also visualized at higher magnifications (Zoom). Scale bars represent 5 μm. The data are representative of at least three independent experiments analyzing at least 50 individual cells per experiment and assay point. (c-d) BMDCs were left untreated or transfected with poly(dG:dC) (2.5 μg/ml) for 2 hours and analyzed by confocal microscopy following immunofluorescence staining with DAPI and antibodies against Rad50 and CARD9. Localization of dsDNA, Rad50, and CARD9 in 100 cells per assay point was determined. (c) Percentages of cells containing Rad50-CARD9 aggregates compared to a diffuse localization of Rad50 and CARD9 are shown. (d) Percentages of WT and Card9-/- BMDCs containing cytosolic dsDNA at 2 hours after dsDNA transfection are demonstrated. (e) BMDCs from WT and Card9-/- mice were transfected with dsDNA, the relative number of cells containing Rad50 aggregates and homogenous Rad50 distribution, respectively, is shown. The data are representative of two independent experiments (c-e).
Supplementary Figure 2 ATM is not involved in dsDNA-induced IL-1b production.
BMDCs from WT and Atm-/- mice were transfected with dsDNA (1 - 4 μg/ml) of different origins for 16 hours or stimulated with LPS + ATP. The IL-1β concentrations in the supernatants were determined. The data are represented as the mean + SEM of triplicates of three independent experiments.
Supplementary Figure 3 CARD9 and Bcl-10 control pro-IL-1β synthesis.
BMDCs from mice of the indicated genotype were transfected with dsDNA (1 - 4 μg/ml) of different origins or stimulated with LPS, CpG, and curdlan plus ATP. The pro-IL-1β concentrations were determined in the cell lysates. The data are represented as the mean + SEM of triplicates. One representative of two independent experiments is shown. ND, not detectable.
Supplementary Figure 4 CARD9 regulates DNA-induced transcription of the genes encoding TNF and IL-6.
WT and Card9-/- BMDCs were transfected with poly(dG:dC) (2.5 μg/ml) for the indicated time and TNF (a) and IL-6 (b) transcript levels were measured by quantitative real-time PCR and normalized to β-actin mRNA levels. The data are shown as the mean + SEM of triplicates. One representative of two independent experiments is shown. **p < 0.01, ***p < 0.001, Student's t-test.
Supplementary Figure 5 cGAMP-STING signaling is independent of CARD9.
BMDCs from WT and Card9-/- (a), or WT and Tmem173-/- (b) mice were transfected with the STING activator cGAMP (4 μg/ml) for 6 hours. As control IFN-β production was induced via TLR9 ligation with CpG. IFN-β levels were measured in the supernatant. The data are represented as the mean + SEM of three (a), or two (b) independent experiments.
Supplementary Figure 6 CARD9 is crucial for RNA virus–induced IL-1β generation.
WT and Card9-/-BMDCs were infected with VSV at MOI 1. LPS, CpG, and curdlan plus ATP were used to investigate CARD9-independent and CARD9-dependent IL-1β production, respectively. The IL-1β concentrations in the supernatants were measured. The data are presented as the mean + SEM. One representative of three independent experiments is shown.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1876 kb)
Rights and permissions
About this article
Cite this article
Roth, S., Rottach, A., Lotz-Havla, A. et al. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1β production. Nat Immunol 15, 538–545 (2014). https://doi.org/10.1038/ni.2888
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2888
This article is cited by
-
Macrophage CARD9 mediates cardiac injury following myocardial infarction through regulation of lipocalin 2 expression
Signal Transduction and Targeted Therapy (2023)
-
Bone Marrow Failure and Immunodeficiency Associated with Human RAD50 Variants
Journal of Clinical Immunology (2023)
-
Card9 protects sepsis by regulating Ripk2-mediated activation of NLRP3 inflammasome in macrophages
Cell Death & Disease (2022)
-
Activation of NF-κB signaling via cytosolic mitochondrial RNA sensing in kerotocytes with mitochondrial DNA common deletion
Scientific Reports (2021)
-
TRIM41 is required to innate antiviral response by polyubiquitinating BCL10 and recruiting NEMO
Signal Transduction and Targeted Therapy (2021)