Abstract
Deletion of the DNA-binding domain of the transcription factor Ikaros generates dominant-negative isoforms that interfere with its activity and correlate with poor prognosis in human precursor B cell acute lymphoblastic leukemia (B-ALL). Here we found that conditional inactivation of the Ikaros DNA-binding domain in early pre-B cells arrested their differentiation at a stage at which integrin-dependent adhesion to niches augmented signaling via mitogen-activated protein kinases, proliferation and self-renewal and attenuated signaling via the pre-B cell signaling complex (pre-BCR) and the differentiation of pre-B cells. Transplantation of polyclonal Ikaros-mutant pre-B cells resulted in long-latency oligoclonal pre-B-ALL, which demonstrates that loss of Ikaros contributes to multistep B cell leukemogenesis. Our results explain how normal pre-B cells transit from a highly proliferative and stroma-dependent phase to a stroma-independent phase during which differentiation is enabled, and suggest potential therapeutic strategies for Ikaros-mutant B-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Monroe, J.G. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat. Rev. Immunol. 6, 283–294 (2006).
Herzog, S., Reth, M. & Jumaa, H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9, 195–205 (2009).
Gauld, S.B. & Cambier, J.C. Src-family kinases in B-cell development and signaling. Oncogene 23, 8001–8006 (2004).
Kitamura, D. et al. A critical role of λ5 protein in B cell development. Cell 69, 823–831 (1992).
Gong, S. & Nussenzweig, M.C. Regulation of an early developmental checkpoint in the B cell pathway by Ig β. Science 272, 411–414 (1996).
Kraus, M. et al. Interference with immunoglobulin (Ig)α immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation modulates or blocks B cell development, depending on the availability of an Igβ cytoplasmic tail. J. Exp. Med. 194, 455–469 (2001).
Pelanda, R., Braun, U., Hobeika, E., Nussenzweig, M.C. & Reth, M. B cell progenitors are arrested in maturation but have intact VDJ recombination in the absence of Ig-α and Ig-β. J. Immunol. 169, 865–872 (2002).
Cheng, A.M. et al. Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378, 303–306 (1995).
Schweighoffer, E., Vanes, L., Mathiot, A., Nakamura, T. & Tybulewicz, V.L. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity 18, 523–533 (2003).
Saijo, K. et al. Essential role of Src-family protein tyrosine kinases in NF-κB activation during B cell development. Nat. Immunol. 4, 274–279 (2003).
Marshall, A.J., Fleming, H.E., Wu, G.E. & Paige, C.J. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J. Immunol. 161, 6038–6045 (1998).
Fleming, H.E. & Paige, C.J. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity 15, 521–531 (2001).
Malin, S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11, 171–179 (2010).
Yasuda, T. et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity 28, 499–508 (2008).
Kersseboom, R. et al. Bruton's tyrosine kinase cooperates with the B cell linker protein SLP-65 as a tumor suppressor in Pre-B cells. J. Exp. Med. 198, 91–98 (2003).
Middendorp, S. et al. Tumor suppressor function of Bruton tyrosine kinase is independent of its catalytic activity. Blood 105, 259–265 (2005).
Wen, R. et al. Essential role of phospholipase Cγ2 in early B-cell development and Myc-mediated lymphomagenesis. Mol. Cell. Biol. 26, 9364–9376 (2006).
Herzog, S. et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 9, 623–631 (2008).
Johnson, K. et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28, 335–345 (2008).
Ochiai, K. et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 13, 300–307 (2012).
Cobaleda, C. & Sanchez-Garcia, I. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. Bioessays 31, 600–609 (2009).
Mullighan, C.G. et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 (2007).
Mullighan, C.G. et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 360, 470–480 (2009).
Iacobucci, I. et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood 112, 3847–3855 (2008).
Iacobucci, I. et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 114, 2159–2167 (2009).
Harvey, R.C. et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 116, 4874–4884 (2010).
Georgopoulos, K. Acute lymphoblastic leukemia–on the wings of IKAROS. N. Engl. J. Med. 360, 524–526 (2009).
Georgopoulos, K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994).
Ng, S.Y., Yoshida, T., Zhang, J. & Georgopoulos, K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507 (2009).
Morgan, B. et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16, 2004–2013 (1997).
Thompson, E.C. et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26, 335–344 (2007).
Sun, L., Liu, A. & Georgopoulos, K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 15, 5358–5369 (1996).
Pelanda, R., Schaal, S., Torres, R.M. & Rajewsky, K. A prematurely expressed Igκ transgene, but not VκJκ gene segment targeted into the Igκ locus, can rescue B cell development in λ5-deficient mice. Immunity 5, 229–239 (1996).
Rolink, A.G., Winkler, T., Melchers, F. & Andersson, J. Precursor B cell receptor-dependent B cell proliferation and differentiation does not require the bone marrow or fetal liver environment. J. Exp. Med. 191, 23–32 (2000).
Kierney, P.C. & Dorshkind, K. B lymphocyte precursors and myeloid progenitors survive in diffusion chamber cultures but B cell differentiation requires close association with stromal cells. Blood 70, 1418–1424 (1987).
Hayashi, S. et al. Stepwise progression of B lineage differentiation supported by interleukin 7 and other stromal cell molecules. J. Exp. Med. 171, 1683–1695 (1990).
Glodek, A.M. et al. Focal adhesion kinase is required for CXCL12-induced chemotactic and pro-adhesive responses in hematopoietic precursor cells. Leukemia 21, 1723–1732 (2007).
Tse, K.W. et al. B cell receptor-induced phosphorylation of Pyk2 and focal adhesion kinase involves integrins and the Rap GTPases and is required for B cell spreading. J. Biol. Chem. 284, 22865–22877 (2009).
Rolink, A., Streb, M., Nishikawa, S. & Melchers, F. The c-kit-encoded tyrosine kinase regulates the proliferation of early pre-B cells. Eur. J. Immunol. 21, 2609–2612 (1991).
Sudo, T. et al. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA 90, 9125–9129 (1993).
Winandy, S., Wu, P. & Georgopoulos, K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83, 289–299 (1995).
Ye, F., Kim, C. & Ginsberg, M.H. Reconstruction of integrin activation. Blood 119, 26–33 (2012).
Wehrle-Haller, B. Structure and function of focal adhesions. Curr. Opin. Cell Biol. 24, 116–124 (2012).
Galbraith, C.G., Yamada, K.M. & Galbraith, J.A. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science 315, 992–995 (2007).
Vicente-Manzanares, M., Choi, C.K. & Horwitz, A.R. Integrins in cell migration–the actin connection. J. Cell Sci. 122, 199–206 (2009).
Smith, A. et al. A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J. Cell Biol. 170, 141–151 (2005).
Choi, C.K. et al. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 (2008).
Heasman, S.J. & Ridley, A.J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 (2008).
Park, S.Y. et al. Focal adhesion kinase regulates the localization and retention of pro-B cells in bone marrow microenvironments. J. Immunol. 190, 1094–1102 (2013).
Waanders, E. et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia 25, 254–258 (2011).
Yoshida, T., Ng, S.Y., Zuniga-Pflucker, J.C. & Georgopoulos, K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7, 382–391 (2006).
Schlissel, M.S., Corcoran, L.M. & Baltimore, D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173, 711–720 (1991).
Fuxa, M. et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18, 411–422 (2004).
Osmond, D.G., Melchers, F. & Paige, C.J. Pre-B cells in mouse bone marrow: in vitro maturation of peanut agglutinin binding B lymphocyte precursors separated from bone marrow by fluorescence-activated cell sorting. J. Immunol. 133, 86–90 (1984).
Bernardi, P., Patel, V.P. & Lodish, H.F. Lymphoid precursor cells adhere to two different sites on fibronectin. J. Cell Biol. 105, 489–498 (1987).
Roumiantsev, S., de Aos, I.E., Varticovski, L., Ilaria, R.L. & Van Etten, R.A. The src homology 2 domain of Bcr/Abl is required for efficient induction of chronic myeloid leukemia-like disease in mice but not for lymphoid leukemogenesis or activation of phosphatidylinositol 3-kinase. Blood 97, 4–13 (2001).
Acknowledgements
We thank B. Czyzewski for mouse husbandry; K. White, J.M. Park, B. Morgan, R. Bakshi, E. Alonzo and J. Seavitt for critical review of the manuscript; R. Bakshi for assistance with statistical analysis; and R. Arya for assistance with confocal microscopy. High-throughput DNA sequencing was done at the Bauer Center for Genomic Research (Harvard University). Supported by the US National Institutes of Allergy and Infectious Diseases (American Recovery and Reinvestment Act supplement to 5R01AI 42254-14) and the National Cancer Institute (5R01CA162092-20 to K.G. and CA090576 to R.A.V.E.).
Author information
Authors and Affiliations
Contributions
I.J., T.Y., N.J., X.Q. and J.Z. did the experiments and edited the manuscript; I.J. and T.Y. created figures; and R.A.V.E. and K.G. supervised the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
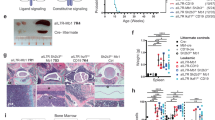
Supplementary Figure 1 Analysis of B-lymphoid differentiation in Ikaros-mutant BM.
a, Schematic representation of B cell differentiation as defined by stage-specific markers. Dotted lines indicate differentiation stages with CD2- or CD19-Cre activity, red lines the differentiation block associated with germline or conditional Ikaros gene mutations, and red arrow the stage from which B-ALL is derived. b-c, Representative flow cytometric analyses of wild-type (WT), IkE5fl/fl CD19-Cre (b) and Ikzf3−/− Ikzf1+/− (c) BM cells as described in Fig. 1d, demonstrating a consistent block at the large pre-B cell stage. IkE5fl/fl CD19-Cre, n=9; Ikzf3−/− Ikzf1+/−, n=3. d, Deletion analysis of the Ikzf1 locus in pro-B cells (CD19+CD43+c-Kit+BP1−) and immature B cells (CD19+IgM+) sorted from BM of IkE5fl/fl CD2-Cre mice.
Supplementary Figure 2 Analysis of B-lymphoid differentiation in immunoglobulin κ-chain−reconstituted Ikaros-mutant pre-B cells.
Flow cytometric analysis of BM B cells from WT, D23, IkE5fl/fl CD2-Cre and IkE5fl/fl CD2-Cre:D23 and intracellular staining for IgM and Igκ in large pre-B cells (CD19+CD43+BP1+).
Supplementary Figure 3 Characterization of adherent and nonadherent wild-type pre-B cells.
a, The mean pro-apoptotic index (percentage of Annexin V+ cells) of WT and IkE5Δ/Δ adherent (left panel) and non-adherent (right panel) pre-B cells propagated on OP9 stroma with 5 ng/ml of supplemental IL-7. Asterisk denotes significant changes in apoptosis between WT and mutant pre-B cells (n=2, *P < 0.05). b, Representative cell cycle profiles of WT adherent and WT non-adherent pre-B cells grown as in Fig. 2a. WT non-adherent pre-B cells were further subdivided according to FSC. The ratio of small vs. large non-adherent WT pre-B in IL-7 cultures increases over time (data not shown). The progressive loss in proliferation in the WT non-adherent pre-B cell phase seen even in the presence of IL-7, suggests a need for stromal contact for maintenance of pre-B cell proliferation. Withdrawal of IL-7 accelerates this process with the ratio of small-non-cycling/large-cycling non-adherent pre-B cells increasing dramatically within 24 hrs (data not shown).
Supplementary Figure 4 Signaling pathways in wild-type and Ikaros-deficient pre-B cells.
a. Schematic representation of signaling pathways operating downstream of pre-BCR and IL-7R and supporting pre-B cell proliferation, survival and differentiation. Signaling effectors assayed for expression and activity in Fig. 4a, b are shown. b, Ca2+ flux (Ca2+ Green/Fura Red) after ionomycin treatment of WT and IkE5Δ/Δ adherent and WT non-adherent pre-B cells, n=2. c, Total Blk expression is shown for WT and IkE5Δ/Δ adherent and non-adherent pre-B cells, with total Akt (T-Akt) as loading control.
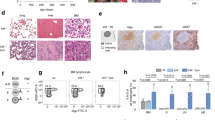
Supplementary Figure 5 Lack of circulating IkE5Δ/Δ pre-B cells and decrease in phosphorylated FAK by FAK inhibitor.
a, Flow cytometric analysis of peripheral blood from wild-type (WT) and IkE5fl/fl CD19-Cre mice for large pre-B (CD19+CD43+) and small pre-B cells (CD19+CD43−); n=2 for each genotype. b, FAK inhibitor treatment reduces p-FAK staining in BM IkE5Δ/Δ pre-B cells, as described in Fig. 6b.
Supplementary Figure 6 Model of pre-BCR, growth factor, and integrin signalling interactions operating during pre-B cell differentiation.
Augmentation of integrin signaling by IkE5Δ/Δ mutation blocks cells in a stromal-dependent, self-renewing and highly proliferative state where they are unable to differentiate, from which B-ALL arises.
Supplementary Figure 7 Clinicopathological characterization of lymphoid tumors from recipients of IkE5Δ/Δ pre-B cells.
a, Immunophenotypic analysis of precursor B-cell acute lymphoblastic leukemia/lymphoma derived from IkE5Δ/Δ pre-B cells demonstrates a similar large pre-B cell surface phenotype (CD19+CD43+BP1+CD2−) to the original transplanted population. b, Analysis of parental WT and IkE5Δ/Δ pre-B cell populations (non-adherent and adherent), showing polyclonal Igh rearrangements similar to that observed in WT splenocytes. The PCR-based D-J rearrangement assay described in Fig. 1f was used to determine clonality. PCR products were probed with a JH-specific probe. c, PCR analysis of V-D-J rearrangements in lymphoid tumors from NSG recipients of IkE5Δ/Δ pre-B cells, as described in Fig. 1f. Forward primers from specific VH regions (558, Q52, 7183) were used in conjunction with a common reverse primer from JH3 (Fig. 1f). Note that lymphoid tumors from mice #391, 393, and 394 (from IkE5Δ/Δ CD19-Cre donor) had monoclonal Igh rearrangement while #392 tumor had clonal rearrangement of both Igh alleles. d, Southern blot analysis of Igh gene rearrangements in tissues of leukemic NSG recipients of IkE5Δ/Δ pre-B cells, as in panel c. The position of two germline (GL) Igh bands (present in control BM myeloid cells, “C”) is denoted by arrowheads. The tissue origin of the sample is indicated (Sp, spleen; LN, lymph node). Common rearrangements between tumors from IkE5Δ/Δ CD19-Cre recipients are indicated by asterisks. Rearrangements in IkE5Δ/Δ CD2-Cre recipients #385 and 386 may not be detected by this probe.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 3978 kb)
Rights and permissions
About this article
Cite this article
Joshi, I., Yoshida, T., Jena, N. et al. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol 15, 294–304 (2014). https://doi.org/10.1038/ni.2821
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2821
This article is cited by
-
IKAROS: from chromatin organization to transcriptional elongation control
Cell Death & Differentiation (2023)
-
Activated interleukin-7 receptor signaling drives B-cell acute lymphoblastic leukemia in mice
Leukemia (2022)
-
A double-negative thymocyte-specific enhancer augments Notch1 signaling to direct early T cell progenitor expansion, lineage restriction and β-selection
Nature Immunology (2022)
-
Fine-tuning Notch1 by the stage-specific enhancer
Nature Immunology (2022)
-
Advances in germline predisposition to acute leukaemias and myeloid neoplasms
Nature Reviews Cancer (2021)