Abstract
Induction of type I interferon is a central event of innate immunity, essential for host defense. Here we report that the transcription factor ELF4 is induced by type I interferon and upregulates interferon expression in a feed-forward loop. ELF4 deficiency leads to reduced interferon production, resulting in enhanced susceptibility to West Nile virus encephalitis in mice. After viral infection, ELF4 is recruited by STING, interacts with and is activated by the MAVS-TBK1 complex, and translocates into the nucleus to bind interferon promoters. Cooperative binding with ELF4 increases the binding affinity of interferon regulatory factors IRF3 and IRF7, which is mediated by EICE elements. Thus, in addition to identifying a regulator of innate immune signaling, we uncovered a role for EICE elements in interferon transactivation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Takeuchi, O. & Akira, S. Innate immunity to virus infection. Immunol. Rev. 227, 75–86 (2009).
Liew, F.Y., Xu, D., Brint, E.K. & O'Neill, L.A. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 (2005).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Seth, R.B., Sun, L., Ea, C.K. & Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005).
Xu, L.G. et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740 (2005).
Barber, G.N. Cytoplasmic DNA innate immune pathways. Immunol. Rev. 243, 99–108 (2011).
Chiu, Y.H., Macmillan, J.B. & Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009).
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010).
Parvatiyar, K. et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161 (2012).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Ishikawa, H. & Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 106, 8653–8658 (2009).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Chen, H. et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147, 436–446 (2011).
Daffis, S., Suthar, M.S., Szretter, K.J., Gale, M. Jr. & Diamond, M.S. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 5, e1000607 (2009).
Yamada, T., Park, C.S., Mamonkin, M. & Lacorazza, H.D. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat. Immunol. 10, 618–626 (2009).
Sashida, G. et al. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol. Cell Biol. 29, 3687–3699 (2009).
Meadows, S.M., Myers, C.T. & Krieg, P.A. Regulation of endothelial cell development by ETS transcription factors. Semin. Cell Dev. Biol. 22, 976–984 (2011).
Sivina, M. et al. The transcription factor E74-like factor controls quiescence of endothelial cells and their resistance to myeloablative treatments in bone marrow. Arterioscler. Thromb. Vasc. Biol. 31, 1185–1191 (2011).
Yao, J.J. et al. Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene 26, 4032–4037 (2007).
Mao, S., Frank, R.C., Zhang, J., Miyazaki, Y. & Nimer, S.D. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell Biol. 19, 3635–3644 (1999).
Gobin, S.J., Biesta, P. & Van den Elsen, P.J. Regulation of human beta 2-microglobulin transactivation in hematopoietic cells. Blood 101, 3058–3064 (2003).
Sauer, J.D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
You, F. et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 10, 1300–1308 (2009).
Suico, M.A. et al. Functional dissection of the ETS transcription factor MEF. Biochim. Biophys. Acta 1577, 113–120 (2002).
Suthar, M.S. et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 6, e1000757 (2010).
Wang, T. et al. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 10, 1366–1373 (2004).
Lacorazza, H.D. et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–449 (2002).
Diamond, M.S., Shrestha, B., Mehlhop, E., Sitati, E. & Engle, M. Innate and adaptive immune responses determine protection against disseminated infection by West Nile encephalitis virus. Viral Immunol. 16, 259–278 (2003).
Vargin, V.V. & Semenov, B.F. Changes of natural killer cell activity in different mouse lines by acute and asymptomatic flavivirus infections. Acta Virol. 30, 303–308 (1986).
Shrestha, B. & Diamond, M.S. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 78, 8312–8321 (2004).
Tough, D.F. Modulation of T-cell function by type I interferon. Immunol. Cell Biol. 90, 492–497 (2012).
Yokoyama, M. et al. Inducible histamine protects mice from P. acnes-primed and LPS-induced hepatitis through H2-receptor stimulation. Gastroenterology 127, 892–902 (2004).
Miyahira, A.K., Shahangian, A., Hwang, S., Sun, R. & Cheng, G. TANK-binding kinase-1 plays an important role during in vitro and in vivo type I IFN responses to DNA virus infections. J. Immunol. 182, 2248–2257 (2009).
Wei, G.H. et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 (2010).
Nolan, G.P. NF-AT-AP-1 and Rel-bZIP: hybrid vigor and binding under the influence. Cell 77, 795–798 (1994).
Bonnefoy, E., Bandu, M.T. & Doly, J. Specific binding of high-mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell Biol. 19, 2803–2816 (1999).
Collett, G.P. & Campbell, F.C. Overexpression of p65/RelA potentiates curcumin-induced apoptosis in HCT116 human colon cancer cells. Carcinogenesis 27, 1285–1291 (2006).
Sato, M. et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13, 539–548 (2000).
Lazear, H.M. et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 9, e1003118 (2013).
Verger, A. & Duterque-Coquillaud, M. When Ets transcription factors meet their partners. BioEssays 24, 362–370 (2002).
Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507 (2012).
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012).
Papachristou, D.J., Batistatou, A., Sykiotis, G.P., Varakis, I. & Papavassiliou, A.G. Activation of the JNK-AP-1 signal transduction pathway is associated with pathogenesis and progression of human osteosarcomas. Bone 32, 364–371 (2003).
Panne, D., Maniatis, T. & Harrison, S.C. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 23, 4384–4393 (2004).
Kong, K.F. et al. Dysregulation of TLR3 impairs the innate immune response to West Nile virus in the elderly. J. Virol. 82, 7613–7623 (2008).
Wang, P. et al. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat. Immunol. 11, 912–919 (2010).
Samuel, M.A. et al. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80, 7009–7019 (2006).
Acknowledgements
We thank S. Nimer (University of Miami) for the Elf4−/− mice, Z. Jiang (Peking University) for the reporter plasmids. This work was supported by the US National Institutes of Health (N01-HHSN272201100019C, AI099625 and AI08992). E.F. and A.I. are funded by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
F.Y. and E.F. designed the study, analyzed the data and wrote or revised the paper. F.Y. performed most experiments. R.L., L.Y. and P.W. analyzed the data. P.W. provided technical support and contributed to animal work. P.W. and L.Y. contributed equally to the second authorship. F.Q. G.Y. and Y.O.Z. provided expertise and contributed to flow cytometry and immunofluorescence experiments and viral infections. L.Y. and W.W. provided technical help. R.M., R.S. and A.I. provided expertise and contributed to experiment with viral infection.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 ELF4 induces IFNβ and interacts with STING.
(a) QRT-PCR (normalized to human β-actin) analysis of IFNβ mRNA after transfecting the 293T cells with the empty vector (EV) or ELF4 at the indicated times the as well as TBK1. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments.There are 2 technical replicates within each experiment. (b) 293T cells were transfected with human IFNβ-Luc or murine IFNβ-Luc and increasing amounts of human ELF4 (hELF4) or murine ELF4 (mELF4). *P < 0.05 and **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (c) Supernatants of the cells in Fig.1h were transfered to 2fTGH-ISRE-Luc cells, and assessed using a luciferase assay. *P < 0.05 and **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (d-h) 293 T cells were transfected with the empty vector (EV), ELF4, STK38 or MAVS and IFNa2-Luc, IFNa4-Luc, IFNa8-Luc, IFNλ3-Luc or IL-22-Luc plasmid, and measured by the luciferase assay. *P < 0.05 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment.
Supplementary Figure 2 C terminus of ELF4 mediates the interaction with STING.
(a) 293T cells were transfected with indicated mutants of ELF4 followed by Western blotting. Data were pooled from 3 independent experiments. (b) Immunoprecipitation (IP), with anti-HA (HA) of proteins from lysates of 293 cells transfected with plasmids encoding hemagglutinin (HA)-tagged STING and Flag-tagged full length or mutant ELF4s, followed by immunoblot analysis (IB) with anti-HA or anti-Flag . Whole lysates (WCL), expression of transfected proteins in whole-cell lysates (without immunoprecipitation). Data were pooled from 3 independent experiments.
Supplementary Figure 3 ELF4 is involved in the antiviral immune signaling.
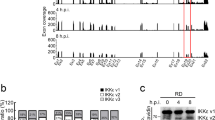
(a&b) 293T cells were transfected with the empty vector (EV), MAVS or increasing amounts of ELF4 and infected with VSV-GFP or WNV, 24 h later, the viral loads were analyzed by microscopy (a) and using a plaque assay (b). **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (c) 293T cells were transfected with empty vector (EV), increasing amounts of Flag-ELF4 or Flag-STK38 and infected with SeV. Lysates were immunoblotted with antibodies to SeV, Flag or GAPDH. Data were pooled from 3 independent experiments. Data were pooled from 3 independent experiments. (d) 293T cells were transfected with the empty vector (EV), scrambled or ELF4 specific siRNA. 48 h later, lysates were immunoblotted using antibodies to ELF4 or GAPDH. (e) 293T cells were transfected with increasing amounts of scrambled or ELF4 specific siRNA and infected with VSV-GFP, 24 hours later, the VSV-GFP was analyzed by FACS. *P < 0.05 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (f) Confirmation of ELF4 knockout by immune blotting. Data were pooled from 3 independent experiments. (g) Wild type, StingGt/Gtor Elf4-/-peritoneal macrophages were infected with VSV-GFP. 24 or 48 h later, the viral loads were analyzed by FACS. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (h) Elf4-/- MEF cells were transfected with wild type ELF4 or the mutant lacking ETS domain. 24 hours later, the cells along with wild type MEF and untransfected cells were infected with VSV-GFP. Data were pooled from 3 independent experiments. (i&j) Development of specific IgG and IgM against WNV. Serum was collected from wild-type or Elf4-/- mice at the indicated days after infection. The levels of specific IgG were determined by incubating serum with adsorbed control or purified WNV E protein. Data are the mean of serum from 8 mice per time point performed in duplicate. * P<0.05 (nonparametric Mann-Whitney analysis).Data were pooled from 3 independent experiments. (k&l) Cxcl10 and IL-6 mRNA (relative to β-actin expression) in whole blood from mice at days 0, 1 and 3 after infection with 200 PFU of WNV subcutaneously. Data were pooled from 3 independent experiments. (m) NK cells and NK1.1+CD3+ NKT cells were sorted from splenocytes of WT mice. 1X106 NK cells and 1X106 NKT cells were injected intravenously (i.v.) into recipient mice. Mortality of age- and sex-matched wild-type (WT; C57BL/6) and Elf4-/-mice inoculated subcutaneously (s.c.; n = 10 mice per group) with 200 PFU of WNV and monitored daily for 15 d. p<0.0001 using the Log-rank test. Data were pooled from 2 independent experiments.(n&o) Proportion of CD4+ T cells and CD8+T cells in spleen of mice at day 0, or day6 after subcutaneous infection with 200 PFU of WNV (s.c.; n = 8 mice per group), assessed by FACS. Data were pooled from 3 independent experiments. No randomization or blinding was used in the animal experiments.
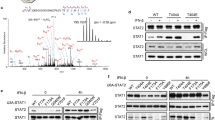
Supplementary Figure 4 ELF4 is involved in TLR-, RLR- and STING-mediated signaling.
(a) 293T cells were transfected with scrambled siRNA, MAVS specific siRNA or ELF4 specific siRNA. 24 h later, cells were infected with SeV or transfected with poly(IC). 24 h later, the according supernatants were transferred to 2fTGH-ISRE-Luc cells. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (b) Wild type, StingGT/GT or Elf4-/- bone marrow derived macrophages (BMDM) were treated with VSV or PRR ligands, LPS, R848, or transfected with poly(dAdT). The TNFα protein concentration in the cell culture medium was measured by ELISA. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (c) Wild type, StingGT/GT or Elf4-/- MEF cells were treated with SeV, EMCV, or HSV-1,. The cellular Cxcl10 transcript abundance was quantified by qRT-PCR (normalized to β-actin expression). *P<0.05 and **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (d) 293T cells were transfected with IFNβ-Luc plasmid increasing amounts of scrambled siRNA. MAVS, STING or TRIF specific siRNA as well as ELF4, MAVS, TRIF or STING. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (e-h). 293T cells were transfected with IFNβ-Luc plasmid or ISRE-Luc plasmid and increasing amounts of scrambled siRNA or ELF4 specific siRNA as well as MAVS (e), TRIF (f) , STING (g) or MyD88 (h). **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment.
Supplementary Figure 5 ELF4 is a new type I interferon regulatory factor.
(a) ELF4 siRNA, MAVS siRNA or scrambled siRNA were transfected into 293T cells. The cells were infected with SeV 36 hours after transfection, with medium (Med) as a control. 16 hours later, the dimerization of IRF3 was detected by native PAGE and immunoblotting. The knockdown efficiency was analyzed by SDS PAGE and immunoblotting. Data were pooled from 3 independent experiments. (b) 293 cells were co-transfected with HA tagged ELF4 and MAVS or TBK1 with or without VSV tagged STING. Cell lysates were immunoprecipitated with Flag and blotted with HA or Flag antibodies. Whole cell lysates (WCL) were blotted with Flag or HA antibodies. Data were pooled from 3 independent experiments. (c) HEK293T cells were treated with medium or SeV. 6 hours later, cell lysates were immunoprecipiated (IP) by anti-ELF4, and immunoblotted (IB) by indicated antibody. Data were pooled from 3 independent experiments. (d) Wild-type or the indicated mutants of ELF4 and IFNβ-Luc were transfected into 293T cells. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (e) In vitro expressed ELF4 and mutantELF4 was incubated with TBK1. The kinase activity of TBK1 is detected by luciferase assay. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (f&g) Wild type and Elf4-/- pDCs were treated with CpG DNA. 8 hours later, the supernatants were transfer to L929-ISRE-Luc cells, followed by luciferase assay (f). The cellular Cxcl10 transcript abundance was quantified by qRT-PCR (g). Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (h&i) Wild-type or the indicated mutants of ELF4 were transfected into 293T cells. Native PAGE and Co-immunoprecipitation were performed at 24 hour after transfection. Data were pooled from 3 independent experiments. (j) Immune fluorescence image of 293T cells to analyze the location of exogenous HA-ELF4 at the indicated time after transfection. Data were pooled from 3 independent experiments. (k) WT or TBK1-/- MEF cells were infected with SeV and the location of ELF4 was determined by microscopy. Data were pooled from 3 independent experiments. (l) HEK293T were transected with wild-type or the indicated mutants and the location of ELF4 was determined by microscopy. Data were pooled from 3 independent experiments. (m) Wild-type or the indicated mutants of ELF4 and IFNβ-Luc were transfected into 293T cells. ****P<0.0001 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (n) There are three GGAA and one ETS-IRF composite element (EICE) in IFNβ promoter. There are four GGAA and one EICE in IFNα2 promoter. There are one GGAA and one EICE in IFNα4 promoter.
Supplementary Figure 6 ELF4 is important for NF-κB– and ISRE-mediated transcription.
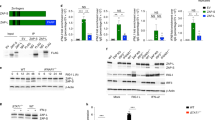
(a) 293T cells were transfected with empty vector (EV), ELF4 or MAVS and IFNβ-Luc, NFκB-Luc or AP1-Luc plasmid. *P<0.05 and ***P<0.001 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (b) Knockdown efficiency of p65 siRNA was analyzed in 293T cells by immunoblotting. Data were pooled from 3 independent experiments. (c&f) 293T cells were co-transfected with p65 or IRF3 and empty vector (EV) or increasing amounts of ELF4 and IFNβ-Luc (c) or NFκB-Luc (f) plasmid. ***P<0.001 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (d) 93T cells were transfected with Flag tagged ELF4 and HA tagged p50, p65 or IRF3. Cell lysates were immunoprecipitated with Flag and blotted with HA or Flag antibodies. WCL were blotted with Flag or HA antibodies. Data were pooled from 3 independent experiments.(e) MEF cells were transfected with scrambled siRNA or p65 specific siRNA. 36 hours later, cells were infected with SeV for 6 hours followed by chromatin immunoprecipitation. The cell lysates were immunoprecipitated by mouse IgG, or anti-ELF4, followed by qRT-PCR to quantify the IFNα2 and IFNα4 promoter DNA. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (g&i) 293T cells were co-transfected with IRF7 and empty vector (EV) or increasing amounts of ELF4 and IFNα2-Luc (g) or ISRE-Luc (i) plasmid. **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (h) 293T cells were co-transfected with IRF3 and empty vector (EV) or increasing amounts of ELF4 and ISRE-Luc plasmid. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (j) In vitro expressed IRF3, p65 or ELF4 was incubated with the biotinylated IFNβ promoter probes followed by EMSA assay. Biotinylated probes were detected by HRP-streptavidin. Free probe (P), Shifted probe (S). Data were pooled from 3 independent experiments.
Supplementary Figure 7 Functional comparison of IRF3, IRF7 and ELF4.
(a) Expression profile of IRF3, IRF7 and ELF4 in mouse analyzed by qRT-PCR(normalized by β-actin). Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment. (b&c) Peritoneal macrophages from Wild-type(WT), Elf4-/- or Irf3-/- were treated with LPS, infected with VSV or HSV-1. QRT-PCR measured the abundance of IFNβ (H) or ISG15 (I). *P<0.05 and **P<0.01 as determined by Student's t-test. Data were pooled from 3 independent experiments. There are 2 technical replicates within each experiment.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–3 (PDF 2349 kb)
Rights and permissions
About this article
Cite this article
You, F., Wang, P., Yang, L. et al. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat Immunol 14, 1237–1246 (2013). https://doi.org/10.1038/ni.2756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2756
This article is cited by
-
“Deficiency in ELF4, X-Linked”: a Monogenic Disease Entity Resembling Behçet’s Syndrome and Inflammatory Bowel Disease
Journal of Clinical Immunology (2024)
-
A Multicenter Cohort Study of Immune Dysregulation Disorders Caused by ELF4 Variants in China
Journal of Clinical Immunology (2023)
-
Intrinsic cardiac adrenergic cells contribute to LPS-induced myocardial dysfunction
Communications Biology (2022)
-
Non-structural proteins of bovine viral diarrhea virus
Virus Genes (2022)
-
Loss of Function Mutation in ELF4 Causes Autoinflammatory and Immunodeficiency Disease in Human
Journal of Clinical Immunology (2022)