Abstract
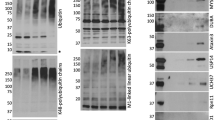
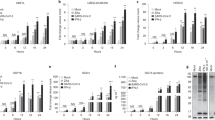
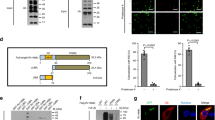
The mammalian immune system has the ability to discriminate between pathogenic microbes and nonpathogenic microbes to control inflammation. Here we investigated the ubiquitination profiles of host proteins after infection of macrophages with a virulent strain of the intracellular bacterium Legionella pneumophila or a nonpathogenic mutant of L. pneumophila. Only infection with pathogenic L. pneumophila resulted in ubiquitination of positive regulators of the metabolic checkpoint kinase mTOR and led to diminished mTOR activity. Detection of pathogen signatures resulted in translational biasing toward proinflammatory cytokines through mTOR-mediated regulation of cap-dependent translation. Thus, there is a pathogen-detection program in macrophages that stimulates protein ubiquitination and the degradation of regulators of mTOR, which suppresses mTOR function and directs a proinflammatory cytokine program.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Newton, K. & Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4, 1–20 (2012).
Vance, R.E., Isberg, R.R. & Portnoy, D.A. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6, 10–21 (2009).
Weintz, G. et al. The phosphoproteome of toll-like receptor-activated macrophages. Mol. Syst. Biol. 6, 371 (2010).
Jiang, X. & Chen, Z.J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12, 35–48 (2011).
Choudhary, C. & Mann, M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 (2010).
Golebiowski, F. et al. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24 (2009).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Vance, R.E. Immunology taught by bacteria. J. Clin. Immunol. 30, 507–511 (2010).
Isberg, R.R., O'Connor, T.J. & Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009).
Sadosky, A.B., Wiater, L.A. & Shuman, H.A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61, 5361–5373 (1993).
Berger, K.H. & Isberg, R.R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 (1993).
Hubber, A. & Roy, C.R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 (2010).
Shin, S. et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 4, e1000220 (2008).
Monroe, K.M., McWhirter, S.M. & Vance, R.E. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 5, e1000665 (2009).
Zamboni, D.S. et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7, 318–325 (2006).
Losick, V.P. & Isberg, R.R. NF-κB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203, 2177–2189 (2006).
Laplante, M. & Sabatini, D.M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Powell, J.D., Pollizzi, K.N., Heikamp, E.B. & Horton, M.R. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 30, 39–68 (2011).
Benveniste, E.N. & Qin, H. Type I interferons as anti-inflammatory mediators. Sci. STKE 2007, pe70 (2007).
Banchereau, J., Pascual, V. & O'Garra, A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nat. Immunol. 13, 925–931 (2012).
Cambronne, E.D. & Roy, C.R. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3, e188 (2007).
Coers, J. et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38, 719–736 (2000).
Fukao, T. et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat. Immunol. 3, 875–881 (2002).
Aksoy, E. et al. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat. Immunol. 13, 1045–1054 (2012).
Weichhart, T. et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29, 565–577 (2008).
Schmidt, E.K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Feldman, M.E. et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38 (2009).
Moerke, N.J. et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128, 257–267 (2007).
Choy, A. et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012).
Fontana, M.F. et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7, e1001289 (2011).
Higgins, D.E., Shastri, N. & Portnoy, D.A. Delivery of protein to the cytosol of macrophages using Escherichia coli K-12. Mol. Microbiol. 31, 1631–1641 (1999).
Thurston, T.L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 (2009).
Creasey, E.A. & Isberg, R.R. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc. Natl. Acad. Sci. USA 109, 3481–3486 (2012).
Andjelković, M. et al. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272, 31515–31524 (1997).
Yang, W.L. et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009).
Chan, C.H. et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149, 1098–1111 (2012).
Svitkin, Y.V. et al. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell Biol. 25, 10556–10565 (2005).
Randow, F. How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy 7, 304–309 (2011).
Tattoli, I. et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012).
Smith, E.M., Finn, S.G., Tee, A.R., Browne, G.J. & Proud, C.G. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 280, 18717–18727 (2005).
Schabbauer, G. et al. Myeloid PTEN promotes inflammation but impairs bactericidal activities during murine pneumococcal pneumonia. J. Immunol. 185, 468–476 (2010).
Cao, W. et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 9, 1157–1164 (2008).
Colina, R. et al. Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008).
Jaramillo, M. et al. Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe 9, 331–341 (2011).
Chakrabarti, S., Liehl, P., Buchon, N. & Lemaitre, B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12, 60–70 (2012).
Ruland, J. Return to homeostasis: downregulation of NF-κB responses. Nat. Immunol. 12, 709–714 (2011).
Anderson, P., Phillips, K., Stoecklin, G. & Kedersha, N. Post-transcriptional regulation of proinflammatory proteins. J. Leukoc. Biol. 76, 42–47 (2004).
Roy, C.R., Berger, K.H. & Isberg, R.R. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28, 663–674 (1998).
Gangloff, Y.G. et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell Biol. 24, 9508–9516 (2004).
Ivanov, S.S. & Roy, C.R. Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell Microbiol. 11, 261–278 (2009).
Acknowledgements
We thank P. Walters (University of California, San Francisco) for the antibody to puromycin; R. Vance (University of California Berkeley) for the L. pneumophila Δ5less strain; P. Kaiser (University of California Irvine) for the biotinylation sequence expression vector; W. Mothes (Yale University) for the HL116 interferon reporter cell line; A.-M. Dragoi for suggestions and manuscript critique; C. Godecke and F. Sewald for technical assistance with flow cytometry; and X. Liu for assistance with analysis of the proteomics data. Supported by the US National Institutes of Health (AI097847 and AI048770 to C.R.).
Author information
Authors and Affiliations
Contributions
S.S.I. designed and did the experiments, interpreted the data and wrote the manuscript; and C.R.R. designed the experiments, interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4 (PDF 8092 kb)
Rights and permissions
About this article
Cite this article
Ivanov, S., Roy, C. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol 14, 1219–1228 (2013). https://doi.org/10.1038/ni.2740
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2740
This article is cited by
-
Effector-triggered immunity and pathogen sensing in metazoans
Nature Microbiology (2019)
-
Outrunning the Red Queen: bystander activation as a means of outpacing innate immune subversion by intracellular pathogens
Cellular & Molecular Immunology (2017)
-
Regnase-1, a rapid response ribonuclease regulating inflammation and stress responses
Cellular & Molecular Immunology (2017)
-
Tribbles ortholog NIPI-3 and bZIP transcription factor CEBP-1 regulate a Caenorhabditis elegans intestinal immune surveillance pathway
BMC Biology (2016)
-
Opposing regulation of the late phase TNF response by mTORC1-IL-10 signaling and hypoxia in human macrophages
Scientific Reports (2016)