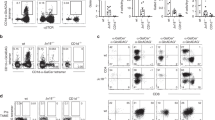
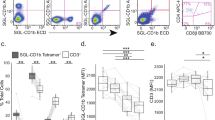
Abstract
The T cell repertoire comprises αβ and γδ T cell lineages. Although it is established how αβ T cell antigen receptors (TCRs) interact with antigen presented by antigen-presenting molecules, this is unknown for γδ TCRs. We describe a population of human Vδ1+ γδ T cells that exhibit autoreactivity to CD1d and provide a molecular basis for how a γδ TCR binds CD1d–α-galactosylceramide (α-GalCer). The γδ TCR docked orthogonally, over the A′ pocket of CD1d, in which the Vδ1-chain, and in particular the germ line–encoded CDR1δ loop, dominated interactions with CD1d. The TCR γ-chain sat peripherally to the interface, with the CDR3γ loop representing the principal determinant for α-GalCer specificity. Accordingly, we provide insight into how a γδ TCR binds specifically to a lipid-loaded antigen-presenting molecule.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Godfrey, D.I., Rossjohn, J. & McCluskey, J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity 28, 304–314 (2008).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012).
Rossjohn, J., Pellicci, D.G., Patel, O., Gapin, L. & Godfrey, D.I. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12, 845–857 (2012).
Brenner, M.B. et al. Identification of a putative second T-cell receptor. Nature 322, 145–149 (1986).
Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of gamma delta T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013).
Schild, H. et al. The nature of major histocompatibility complex recognition by gamma delta T cells. Cell 76, 29–37 (1994).
Crowley, M.P. et al. A population of murine gammadelta T cells that recognize an inducible MHC class Ib molecule. Science 287, 314–316 (2000).
Wingren, C., Crowley, M.P., Degano, M., Chien, Y. & Wilson, I.A. Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold. Science 287, 310–314 (2000).
Willcox, C.R. et al. Cytomegalovirus and tumor stress surveillance by binding of a human gammadelta T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 13, 872–879 (2012).
Constant, P. et al. Stimulation of human gamma delta T cells by nonpeptidic mycobacterial ligands. Science 264, 267–270 (1994).
Vavassori, S. et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human [gamma][delta] T cells. Nat. Immunol. 14, 908–916 (2013).
Russano, A.M. et al. Recognition of pollen-derived phosphatidyl-ethanolamine by human CD1d-restricted gamma delta T cells. J. Allergy Clin. Immunol. 117, 1178–1184 (2006).
Dieude, M. et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J. Immunol. 186, 4771–4781 (2011).
Bai, L. et al. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur. J. Immunol. 42, 2505–2510 (2012).
Godfrey, D.I. & Rossjohn, J. New ways to turn on NKT cells. J. Exp. Med. 208, 1121–1125 (2011).
Girardi, E. et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat. Immunol. 13, 851–856 (2012).
Patel, O. et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat. Immunol. 13, 857–863 (2012).
Xu, B. et al. Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA. Proc. Natl. Acad. Sci. USA 108, 2414–2419 (2011).
Allison, T.J., Winter, C.C., Fournie, J.J., Bonneville, M. & Garboczi, D.N. Structure of a human gammadelta T-cell antigen receptor. Nature 411, 820–824 (2001).
Li, H. et al. Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor. Nature 391, 502–506 (1998).
Adams, E.J., Chien, Y.H. & Garcia, K.C. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 308, 227–231 (2005).
Adams, E.J., Strop, P., Shin, S., Chien, Y.H. & Garcia, K.C. An autonomous CDR3delta is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by gammadelta T cells. Nat. Immunol. 9, 777–784 (2008).
Wun, K.S. et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J. Exp. Med. 205, 939–949 (2008).
Kjer-Nielsen, L. et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J. Exp. Med. 203, 661–673 (2006).
Borg, N.A. et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007).
Mallevaey, T. et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34, 315–326 (2011).
Matulis, G. et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 8, e1000402 (2010).
Pellicci, D.G. et al. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat. Immunol. 12, 827–833 (2011).
Garcia, K.C. et al. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279, 1166–1172 (1998).
Koch, M. et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 6, 819–826 (2005).
Godfrey, D.I., McCluskey, J. & Rossjohn, J. CD1d antigen presentation: treats for NKT cells. Nat. Immunol. 6, 754–756 (2005).
Russano, A.M. et al. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gamma delta+ T lymphocytes. J. Immunol. 178, 3620–3626 (2007).
Cerundolo, V., Silk, J.D., Masri, S.H. & Salio, M. Harnessing invariant NKT cells in vaccination strategies. Nat. Rev. Immunol. 9, 28–38 (2009).
Wieland Brown, L.C. et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 11, e1001610 (2013).
Brennan, P.J. et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat. Immunol. 12, 1202–1211 (2011).
Brigl, M. et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J. Exp. Med. 208, 1163–1177 (2011).
Nick, S. et al. T cell receptor gamma delta repertoire is skewed in cerebrospinal fluid of multiple sclerosis patients: molecular and functional analyses of antigen-reactive gamma delta clones. Eur. J. Immunol. 25, 355–363 (1995).
Matsuda, J.L. et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192, 741–754 (2000).
Kedzierska, K., Turner, S.J. & Doherty, P.C. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc. Natl. Acad. Sci. USA 101, 4942–4947 (2004).
Szymczak, A.L. et al. Correction of multi-gene deficiency in vivo using a single ′self-cleaving′ 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004).
Heemskerk, M.H. et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood 102, 3530–3540 (2003).
Shima, E.A. et al. Gene encoding the alpha chain of the T-cell receptor is moved immediately downstream of c-myc in a chromosomal 8;14 translocation in a cell line from a human T-cell leukemia. Proc. Natl. Acad. Sci. USA 83, 3439–3443 (1986).
Aricescu, A.R., Lu, W. & Jones, E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 (2006).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Holst, M., Baker, N.A. & Wang, F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I. Algorithms and examples. J. Comput. Chem. 21, 1319–1342 (2000).
Acknowledgements
We thank the staff at the MX1 and MX2 beamlines of the Australian Synchrotron for assistance with data collection, M. Sandoval, M. Ciula, D. Littler, R. Berry, J. Waddington, P. Neeson and D. Ritchie for technical assistance, provision of reagents and/or advice, staff from the flow cytometry facilities in the Melbourne Brain Centre and the Department of Microbiology and Immunology at The University of Melbourne and D. Maksel from the Macromolecular Crystallization Facility at Monash University for technical assistance, P. Savage (Brigham Young University) for providing PBS44 glycolipid, G. Besra (University of Birmingham, UK) and S. Porcelli (Albert Einstein College of Medicine) for providing α-glucosylceramide and the α-GalCer analog OCH, and D. Vignali (St. Jude Children's Research Hospital) and S. Turner (The University of Melbourne) for providing pMIG expression vector. This work was supported by the Australian Research Council and the National Health and Medical Research Council of Australia (NHMRC). N.A.G. is supported by a Leukaemia Foundation of Australia Postgraduate Scholarship; T.B. is supported by a Pfizer Australia Fellowship; S.G. and O.P. are supported by Australian Research Council Future Fellowships; D.G.P. is supported by an NHMRC Peter Doherty Fellowship; J.R. is supported by an NHMRC Australia Fellowship; D.I.G. is supported by an NHMRC Senior Principal Research Fellowship.
Author information
Authors and Affiliations
Contributions
A.P.U. identified and performed cellular and molecular characterization of γδ T cells, and produced TCR protein complexes for crystallographic studies. J.L.N., A.P.U. and S.G. solved the crystal structures and performed structural analysis. T.B. undertook SPR investigations. O.P. designed and generated human CD1d-BirA constructs. R.T.L. generated C1R-CD1d transductants. A.P.U., D.G.P., N.A.G., K.G.M. and R.T.L. performed cell-based experiments, including glycolipid specificity and functional studies. J.R. and D.I.G. were joint senior authors: co-led the investigation, devised the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1-3 and Supplementary Figures 1-4 (PDF 1326 kb)
Rights and permissions
About this article
Cite this article
Uldrich, A., Le Nours, J., Pellicci, D. et al. CD1d-lipid antigen recognition by the γδ TCR. Nat Immunol 14, 1137–1145 (2013). https://doi.org/10.1038/ni.2713
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2713
This article is cited by
-
The emerging roles of γδ T cells in cancer immunotherapy
Nature Reviews Clinical Oncology (2023)
-
γδ T cells: origin and fate, subsets, diseases and immunotherapy
Signal Transduction and Targeted Therapy (2023)
-
CD1-mediated immune responses in mucosal tissues: molecular mechanisms underlying lipid antigen presentation system
Experimental & Molecular Medicine (2023)
-
Expression, Purification, and Crystallization of the Vγ9Vδ2 T-cell Receptor Recognizing Protein/Peptide Antigens
The Protein Journal (2023)
-
Chimeric antigen receptor- and natural killer cell receptor-engineered innate killer cells in cancer immunotherapy
Cellular & Molecular Immunology (2021)