A third population of human T lymphocytes that express αβ T cell antigen receptors with restricted α-chain diversity has been identified. These cells recognize the lipid glucose monomycolate from Mycobacterium tuberculosis presented by CD1b.
Gene rearrangements that create an enormous amount of diversity in the variable regions of antigen receptors are undoubtedly protective for the host, but there are also expanded populations of lymphocytes with very limited diversity of their antigen receptors. In this issue of Nature Immunology, Van Rhijn et al. describe a previously unknown population of human T lymphocytes with highly restricted diversity of the T cell antigen receptor (TCR) α-chain1. These TCRs recognize the lipid glucose monomycolate from Mycobacterium tuberculosis when it is presented by the major histocompatibility complex (MHC) class I–like antigen-presenting molecule CD1b. Therefore, they call these 'germline-encoded, mycoyl-reactive' (GEM) TCRs.
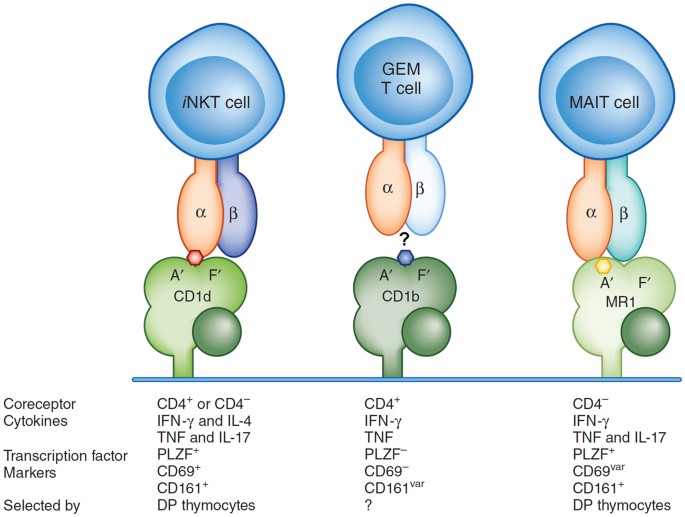
Two populations of T lymphocytes that express αβ TCRs with restricted α-chain diversity are already known. One of these is invariant natural killer T cells (iNKT cells), and the other is mucosa-associated invariant T cells (MAIT cells)2 (Fig. 1). Both of these populations are conserved in mammals, and they each have a different invariant TCR α-chain. In humans, it is composed of α-chain variable region 24 and α-chain joining region 18 (Vα24-Jα18; also known as TRAV10-TRAJ18) for iNKT cells and Vα7.2-Jα33 (TRAV1-2 TRAJ33) for MAIT cells. In each case these α-chains are coexpressed with β-chains that are restricted in Vβ-segment use but not in the diversity of the complementarity-determining region 3 β-chain (CDR3β). The iNKT cells recognize various glycolipids and phospholipids presented by CD1d, a nonpolymorphic MHC class I–like antigen-presenting molecule2. The MAIT cells recognize pterins (microbial riboflavin derivatives) presented by MR1, which is also nonpolymorphic and is the most highly conserved MHC class I–like protein3. Therefore, the expression of an invariant TCRα repertoire is correlated with the recognition of an antigen-presenting molecule with limited polymorphism. These two cell populations have specialized functions in innate-like effector responses, which are very rapid, are generally not accompanied by massive clonal expansion and do not have an obvious memory component. Such T cell populations may be the result of convergent evolution in which some T lymphocytes take on the functional features of cells of the innate immune system.
The known contributions (iNKT cell and MAIT cell) and possible contributions (GEM T cell) of the α- and β-chains to antigen recognition and orientation of the TCR over the antigen-presenting molecule are shown here. The properties listed below emphasize results from studies of human iNKT cells and MAIT cells, except for the identification of the selecting cell type in the thymus, which is from experiments with mice. IFN-γ, interferon-γ; IL, interleukin; TNF, tumor-necrosis factor; superscript 'var', variable expression; DP, double-positive (CD4+CD8+).
Deductively, there is no reason that antigen recognition by host-protective T lymphocytes should be confined only to peptides, and according to the same rationale, there is no reason that T lymphocytes specific for lipid antigens should be confined to a limited TCR repertoire. In fact, there are CD1d-reactive cells with diverse TCRs, sometimes called 'diverse' or 'type II' NKT cells. Not surprisingly, such cells have various specificities that differ from those of type I NKT cells. The evidence suggests, however, that type II NKT cells also have innate-like properties4.
The immune system is not designed for simplicity, and the recognition of lipid antigens is no exception. Although mice and rats studied in the laboratory have only a single CD1 isotype (CD1d; group 2), most mammals have additional CD1 proteins, collectively known as 'group 1 CD1 molecules'. Group 1 includes the isoforms CD1a, CD1b and CD1c, with limited polymorphism. In fact, the recognition of lipid antigens by T cells was first described with the discovery of T cell lines reactive to mycolic acid, a long-chain mycobacterial lipid, presented by CD1b5. Subsequent work has shown that CD1a and CD1c also present various mycobacterial lipids and that the reactive T cells have diverse TCRs and patterns of coreceptor expression and produce cytokines such as interferon-γ6. Those data led to the suggestion that T lymphocytes reactive to group 1 CD1 molecules constitute a branch of adaptive immunity important for protection from mycobacterial infection, in contrast to CD1d-reactive iNKT cells, with their innate-like characteristics. However, progress in understanding T cells reactive to group 1 CD1 molecules has been hampered by several factors. One has been the absence, until recently7, of a mouse experimental model. A second issue, also addressed recently, has been the lack of tetramers of group 1 CD1. Therefore, most research studying group 1 CD1 proteins has used a limited number of long-term cell lines and clones that may not be representative of natural populations. Interestingly, reactivity mediated by group 1 CD1 molecules to lipids from microbes other than mycobacteria has rarely been reported. It is uncertain if this reflects the functioning of a highly specialized antigen-recognition system for mycobacteria selected by evolution or if this simply reflects the interests of the relatively few laboratories that have been able to explore this system.
Van Rhijn et al. use tetramer-based selection of cells from patients with tuberculosis without extensive population expansion in vitro to show that lymphocytes reactive to glucosylated mycolic acid presented by CD1b have a limited TCR diversity. These GEM TCR–expressing T cells ('GEM T cells') express TRAV1-2, the same Vα segment expressed by MAIT cells, but rearranged to TRAJ9, which differs from the MAIT Jα segment. They use mainly TRBV6-2, also found in MAIT cells. Notably, CDR3 of the α-chain is highly restricted in diversity. It is conserved in length, and although two amino acids are variable because of nontemplated additions, the structures of the iNKT cell and MAIT cell TCRs would suggest those amino acids do not directly contact the antigen. GEM T cells are CD4+, and some express the NK cell receptor CD161, which is also expressed by most iNKT cells and MAIT cells, and they are able to secrete interferon-γ and tumor-necrosis factor, cytokines that stimulate antimicrobial responses. Sorting of TRAV1-2-expressing peripheral blood cells from blood-bank donors identifies the presence of CD1b-restricted cells and the GEM sequence motif in healthy people as well. Therefore, the parallels between GEM T cells and type I or iNKT cells are indeed noteworthy. The authors also find a population of peripheral lymphocytes with more diverse TCRs that react to tetramers of CD1b loaded with glucose monomycolate, but these have diminished tetramer staining and, therefore, lower apparent affinity.
An interesting feature of the GEM TCR is that the β-chain apparently regulates antigen specificity between mycolic acid and glucose monomycolate, possibly by affecting the conformation of the conserved CDR3α loop. Also of note, the protein sequence and structure of the TCR reported here shows that GEM TCRs have more similarity with the TCRs of iNKT cells, although they share TRAV1-2 use with the TCRs of MAIT cells. CDR1α and CDR3α of the GEM TCR have residues similar or identical those of the iNKT TCR in positions used by the latter to bind to CD1d and/or the antigen. Because superimposition of the two TCRs shows similar orientations of all six CDR loops, this raises the possibility that GEM TCRs can bind in an orientation similar to that of iNKT cell TCRs above the F′ pocket of CD1b. In contrast, the TCRs of MAIT cells bind above MR1 in an orientation similar to that of a sulfatide-reactive type II NKT TCR that is perpendicular and above the A′ pocket. Therefore, there are apparently two different binding orientations of semi-invariant TCRs above their antigen-presenting molecules8. It remains to be determined if GEM TCRs bind above CD1b in a manner similar to that used by the TCRs of iNKT cells or MAIT cells or if they assume a different orientation altogether.
Despite such similarities, it is unknown if GEM T cells constitute a population of innate-like T lymphocytes, truly similar to iNKT cells and MAIT cells. The iNKT cells and MAIT cells are positively selected by CD4+CD8+ double-positive thymocytes rather than by the cortical epithelial cells that select mainstream T cells9. Their unique developmental program imprints their innate-like properties on them, and this is related to their expression of the transcription factor PLZF (Zbtb16), which is expressed by both iNKT cells and MAIT cells in humans but only by iNKT cells in mice10. GEM T cells do not express PLZF or have high expression of the activation marker CD69 and therefore they not only lack the transcription factor responsible for the effector phenotype of iNKT cells and MAIT cells but also lack the constitutively activated phenotype of those cells. Deep sequencing of samples from two blood-bank donors also indicates that the frequency of GEM T cells is approximately 2 in 1 × 105 cells, which is much less than the frequency of MAIT cells and also less than that of iNKT cells in most people. Therefore, GEM T cells may be 'mainstream', positively selected cells that are truly part of the adaptive immune system that have undergone population expansion in response to antigenic exposure.
If GEM T cells essentially are lipid-reactive, 'mainstream' T lymphocytes, then the question remains of why the human immune system has come up with such a prevalent TCR solution to the problem of the recognition of glucose monomycolate plus CD1b. Perhaps the nonpolymorphic nature of CD1b is part of the answer, as well as the widespread exposure of humans to nonpathogenic mycobacteria with similar antigens. Regardless of the nature of their origin, it will be interesting to determine if populations of GEM T cells expand during infection with other bacteria, such as Mycobacterium leprae. Whether tracking GEM T cells could be useful for the diagnosis of tuberculosis and if stimulation of these cells could be an important target for vaccine development needs to be investigated in the future. Therefore, regardless of their degree of similarity to the innate-like T cells, GEM T cells are truly a 'diamond in the rough'.
References
Van Rhijn, I. et al. Nat. Immunol. 14, 706–713 (2013).
Treiner, E. & Lantz, O. Curr. Opin. Immunol. 18, 519–526 (2006).
Kjer-Nielsen, L. et al. Nature 491, 717–723 (2012).
Sköld, M., Faizunnessa, N.N., Wang, C.R. & Cardell, S. J. Immunol. 165, 168–174 (2000).
Beckman, E.M. et al. Nature 372, 691–694 (1994).
Brigl, M. & Brenner, M.B. Annu. Rev. Immunol. 22, 817–890 (2004).
Felio, K. et al. J. Exp. Med. 206, 2497–2509 (2009).
López-Sagaseta, J. et al. Proc. Natl. Acad. Sci. USA 110, E1771–E1778 (2013).
Gapin, L. PLoS Biol. 7, e70 (2009).
Constantinides, M.G. & Bendelac, A. Curr. Opin. Immunol. 25, 161–167 (2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kronenberg, M., Zajonc, D. A 'GEM' of a cell. Nat Immunol 14, 694–695 (2013). https://doi.org/10.1038/ni.2644
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2644