Abstract
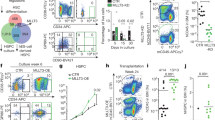
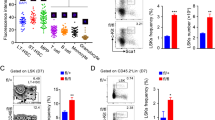
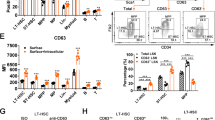
How hematopoietic stem cells (HSCs) coordinate the regulation of opposing cellular mechanisms such as self-renewal and differentiation commitment remains unclear. Here we identified the transcription factor and chromatin remodeler Satb1 as a critical regulator of HSC fate. HSCs lacking Satb1 had defective self-renewal, were less quiescent and showed accelerated lineage commitment, which resulted in progressive depletion of functional HSCs. The enhanced commitment was caused by less symmetric self-renewal and more symmetric differentiation divisions of Satb1-deficient HSCs. Satb1 simultaneously repressed sets of genes encoding molecules involved in HSC activation and cellular polarity, including Numb and Myc, which encode two key factors for the specification of stem-cell fate. Thus, Satb1 is a regulator that promotes HSC quiescence and represses lineage commitment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Till, J.E. & McCulloch, E.A. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 175, 145–149 (1961).
Becker, A.J., McCulloch, E.A. & Till, J.E. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454 (1963).
Spangrude, G.J., Heimfeld, S. & Weissman, I.L. Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62 (1988).
Seita, J. & Weissman, I.L. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 640–653 (2010).
Zeng, H., Yucel, R., Kosan, C., Klein-Hitpass, L. & Moroy, T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23, 4116–4125 (2004).
Perry, J.M. & Li, L. Self-renewal versus transformation: Fbxw7 deletion leads to stem cell activation and leukemogenesis. Genes Dev. 22, 1107–1109 (2008).
Antonchuk, J., Sauvageau, G. & Humphries, R.K. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109, 39–45 (2002).
Chang, K.C. et al. Interplay between the transcription factor Zif and aPKC regulates neuroblast polarity and self-renewal. Dev. Cell 19, 778–785 (2010).
Scheel, C. et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940 (2011).
Bröske, A.M. et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 41, 1207–1215 (2009).
Sun, Y., Hu, J., Zhou, L., Pollard, S.M. & Smith, A. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J. Cell Sci. 124, 1867–1877 (2011).
Alvarez, J.D. et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14, 521–535 (2000).
Yasui, D., Miyano, M., Cai, S., Varga-Weisz, P. & Kohwi-Shigematsu, T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002).
Beyer, M. et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12, 898–907 (2011).
Steidl, U. et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 117, 2611–2620 (2007).
Savarese, F. et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 23, 2625–2638 (2009).
Han, H.J., Russo, J., Kohwi, Y. & Kohwi-Shigematsu, T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193 (2008).
Lakshminarayana Reddy, C.N., Vyjayanti, V.N., Notani, D., Galande, S. & Kotamraju, S. Down-regulation of the global regulator SATB1 by statins in COLO205 colon cancer cells. Mol. Med. Rep. 3, 857–861 (2010).
Selinger, C.I. et al. Loss of special AT-rich binding protein 1 expression is a marker of poor survival in lung cancer. J. Thorac. Oncol. 6, 1179–1189 (2011).
Cai, S., Han, H.J. & Kohwi-Shigematsu, T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51 (2003).
Cai, S., Lee, C.C. & Kohwi-Shigematsu, T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288 (2006).
Challen, G.A., Boles, N., Lin, K.K. & Goodell, M.A. Mouse hematopoietic stem cell identification and analysis. Cytometry A 75, 14–24 (2009).
Matsumoto, A. et al. P57 is Required for Quiescence and Maintenance of Adult Hematopoietic Stem Cells. Cell Stem Cell 9, 262–271 (2011).
Passegué, E., Wagers, A.J., Giuriato, S., Anderson, W.C. & Weissman, I.L. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202, 1599–1611 (2005).
Takizawa, H., Regoes, R.R., Boddupalli, C.S., Bonhoeffer, S. & Manz, M.G. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208, 273–284 (2011).
Wilson, A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008).
Morita, Y., Ema, H. & Nakauchi, H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 207, 1173–1182 (2010).
Kharas, M.G. et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 (2010).
Wu, M. et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554 (2007).
Rhyu, M.S., Jan, L.Y. & Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994).
Spana, E.P. & Doe, C.Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996).
Rebeiz, M., Miller, S.W. & Posakony, J.W. Notch regulates numb: integration of conditional and autonomous cell fate specification. Development 138, 215–225 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Zeller, K.I. et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA 103, 17834–17839 (2006).
Huang, M.J., Cheng, Y.C., Liu, C.R., Lin, S. & Liu, H.E. A small-molecule c-Myc inhibitor, 10058–F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 34, 1480–1489 (2006).
Delmore, J.E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Pinto do, O.P., Kolterud, A. & Carlsson, L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17, 5744–5756 (1998).
Mahmoudi, T. et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 28, 3329–3340 (2009).
Fazzio, T.G., Huff, J.T. & Panning, B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162–174 (2008).
Miyazaki, M. et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12, 992–1001 (2011).
Wontakal, S.N. et al. A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet. 7, e1001392.1–15 (2011).
Nerlov, C. & Graf, T.P.U. 1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 (1998).
Orford, K.W. & Scadden, D.T. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9, 115–128 (2008).
Cheng, T. et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000).
Massagué, J. G1 cell-cycle control and cancer. Nature 432, 298–306 (2004).
Xi, R. & Xie, T. Stem cell self-renewal controlled by chromatin remodeling factors. Science 310, 1487–1489 (2005).
Reavie, L. et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat. Immunol. 11, 207–215 (2010).
Santaguida, M. et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell 15, 341–352 (2009).
Klinakis, A. et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473, 230–233 (2011).
Li, Q.Q. et al. Overexpression and involvement of special AT-rich sequence binding protein 1 in multidrug resistance in human breast carcinoma cells. Cancer Sci. 101, 80–86 (2010).
Kossenkov, A.V. & Ochs, M.F. Matrix factorization for recovery of biological processes from microarray data. Methods Enzymol. 467, 59–77 (2009).
Maggio-Price, L., Wolf, N.S., Priestley, G.V., Pietrzyk, M.E. & Bernstein, S.E. Evaluation of stem cell reserve using serial bone marrow transplantation and competitive repopulation in a murine model of chronic hemolytic anemia. Exp. Hematol. 16, 653–659 (1988).
Szilvassy, S.J., Nicolini, F.E., Eaves, C.J. & Miller, C.L. Quantitation of murine and human hematopoietic stem cells by limiting-dilution analysis in competitively repopulated hosts. Methods Mol. Med. 63, 167–187 (2002).
Lyons, A.B. Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J. Immunol. Methods 243, 147–154 (2000).
Akalin, A. et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 8, e1002781 (2012).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
We thank T. Kohwi-Shigematsu (Lawrence Berkeley National Laboratory) for Satb1−/− mice; J. Bradner (Dana-Farber Cancer Institute, Harvard Medical School) for JQ1; T. Kohwi-Shigematsu, E. Passegué, M. Alberich-Jordá and the members of the Steidl laboratory for discussions and suggestions; G. Simkin, and S. Narayanagari of the Einstein Human Stem Cell FACS and Xenotransplantation Facility; and P. Schultes, C. Sheridan and the Epigenomics Core Facility of Weill Cornell Medical College for technical assistance. Supported by the American Cancer Society (121366-PF-12-89-01-TBG to B.W.), the Sass Foundation (F.G.B.), the National Cancer Institute (1K08CA169055-01 to F.G.B. and R00CA131503 to U.S.), the US National Institutes of Health (F31CA162770 to U.C.O.-O. and F30HL117545 to A.P.) and New York State Stem Cell Science (C024306, C026416 and C028116 to U.S., and C024172 to E. Bouhassira). U.S. is the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
Author information
Authors and Affiliations
Contributions
B.W. and U.S. designed the study and experiments; B.W., T.O.V., F.G.-B., T.D.B., J.M. and T.I.T. did experiments; B.W., T.O.V., J.M., B.B., F.G.-B., A.P., L.B., U.C.O.-O., R.F.S., T.I.T., M.R., A.V., M.E.F., A.M. and U.S. interpreted experiments; B.B. and L.B. did statistical analysis of microarray data; B.B. and F.G.-B. did statistical analysis of ERRBS data; and B.W. and U.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Tables 1–6 (PDF 1133 kb)
Rights and permissions
About this article
Cite this article
Will, B., Vogler, T., Bartholdy, B. et al. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat Immunol 14, 437–445 (2013). https://doi.org/10.1038/ni.2572
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2572
This article is cited by
-
A dual function for the chromatin organizer Special A-T rich Binding Protein 1 in B-lineage cells
Cellular & Molecular Immunology (2023)
-
Ppm1d truncating mutations promote the development of genotoxic stress-induced AML
Leukemia (2023)
-
Cell-intrinsic factors governing quiescence vis-à-vis activation of adult hematopoietic stem cells
Molecular and Cellular Biochemistry (2023)
-
CD63 acts as a functional marker in maintaining hematopoietic stem cell quiescence through supporting TGFβ signaling in mice
Cell Death & Differentiation (2022)
-
Chromatin organizer SATB1 controls the cell identity of CD4+ CD8+ double-positive thymocytes by regulating the activity of super-enhancers
Nature Communications (2022)