Abstract
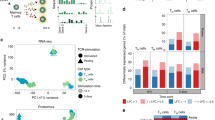
After infection, many factors coordinate the population expansion and differentiation of CD8+ effector and memory T cells. Using data of unparalleled breadth from the Immunological Genome Project, we analyzed the CD8+ T cell transcriptome throughout infection to establish gene-expression signatures and identify putative transcriptional regulators. Notably, we found that the expression of key gene signatures can be used to predict the memory-precursor potential of CD8+ effector cells. Long-lived memory CD8+ cells ultimately expressed a small subset of genes shared by natural killer T and γδ T cells. Although distinct inflammatory milieu and T cell precursor frequencies influenced the differentiation of CD8+ effector and memory populations, core transcriptional signatures were regulated similarly, whether polyclonal or transgenic, and whether responding to bacterial or viral model pathogens. Our results provide insights into the transcriptional regulation that influence memory formation and CD8+ T cell immunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Haining, W.N. et al. Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells. J. Immunol. 181, 1859–1868 (2008).
Kaech, S.M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Sarkar, S. et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008).
Wirth, T.C. et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity 33, 128–140 (2010).
Kaech, S.M. & Ahmed, R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2, 415–422 (2001).
Busch, D.H., Kerksiek, K.M. & Pamer, E.G. Differing roles of inflammation and antigen in T cell proliferation and memory generation. J. Immunol. 164, 4063–4070 (2000).
Badovinac, V.P., Haring, J.S. & Harty, J.T. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8+ T cell response to infection. Immunity 26, 827–841 (2007).
Haluszczak, C. et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206, 435–448 (2009).
Yamada, T., Park, C.S., Mamonkin, M. & Lacorazza, H.D. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat. Immunol. 10, 618–626 (2009).
Beuneu, H. et al. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity 33, 412–423 (2010).
Grayson, J.M., Zajac, A.J., Altman, J.D. & Ahmed, R. Cutting edge: increased expression of Bcl-2 in antigen-specific memory CD8+ T cells. J. Immunol. 164, 3950–3954 (2000).
Gründemann, C. et al. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J. Immunol. 176, 1311–1315 (2006).
Dietz, S.B., Whitaker-Menezes, D. & Lessin, S.R. The role of αEβ7 integrin (CD103) and E-cadherin in epidermotropism in cutaneous T-cell lymphoma. J. Cutan. Pathol. 23, 312–318 (1996).
Pearce, E.L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
van der Windt, G.J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Gautier, E.L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Joshi, N.S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Kaech, S.M. et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003).
Schluns, K.S., Kieper, W.C., Jameson, S.C. & Lefrancois, L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 (2000).
Huster, K.M. et al. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA 101, 5610–5615 (2004).
Yang, C.Y. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011).
Cannarile, M.A. et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7, 1317–1325 (2006).
D'Cruz, L.M., Lind, K.C., Wu, B.B., Fujimoto, J.K. & Goldrath, A.W. Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8+ T-cell immune response. Eur. J. Immunol. 8, 2031–2041 (2012).
Harty, J.T. & Badovinac, V.P. Shaping and reshaping CD8+ T-cell memory. Nat. Rev. Immunol. 8, 107–119 (2008).
Obar, J.J., Khanna, K.M. & Lefrancois, L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28, 859–869 (2008).
Obar, J.J. et al. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J. Immunol. 187, 4967–4978 (2011).
Marzo, A.L. et al. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6, 793–799 (2005).
Agarwal, P. et al. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J. Immunol. 183, 1695–1704 (2009).
Sacerdote, P., Massi, P., Panerai, A.E. & Parolaro, D. In vivo and in vitro treatment with the synthetic cannabinoid CP55, 940 decreases the in vitro migration of macrophages in the rat: involvement of both CB1 and CB2 receptors. J. Neuroimmunol. 109, 155–163 (2000).
Geserick, P., Kaiser, F., Klemm, U., Kaufmann, S.H. & Zerrahn, J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 16, 1535–1548 (2004).
Kallies, A. Distinct regulation of effector and memory T-cell differentiation. Immunol. Cell Biol. 86, 325–332 (2008).
Sundrud, M.S. & Rao, A. Regulation of T helper 17 differentiation by orphan nuclear receptors: it's not just RORγt anymore. Immunity 28, 5–7 (2008).
Acknowledgements
We thank eBioscience, Affymetrix and Expression Analysis for support of the ImmGen Project. Supported by the US National Institutes of Health (AI072117 and AI067545 to A.W.G.; T32 AI060536 to J.A.B.; PN2 EY016586 to D.A.B. and M.L.D.; P30 CA016087 for cell sorting; and R24 AI072073 (National Institute of Allergy and Infectious Diseases) to the ImmGen Consortium), the Pew Scholars program (A.W.G.) and the Cancer Research Institute (A.W.G. and V.M.).
Author information
Authors and Affiliations
Consortia
Contributions
J.A.B. did experiments, designed studies, analyzed data and wrote the manuscript; J.K. sorted cell subsets and analyzed data; A.D. analyzed data and edited the manuscript; D.A.B., V.M. and M.L.D. designed and did early infection experiments, analyzed data and contributed to writing the manuscript; E.Y. sorted cell subsets; and A.W.G. designed studies, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations appears at the end of the paper.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Tables 2–4 and Note (PDF 3722 kb)
Supplementary Table 1
Gene expression values for gene lists defined by clusters I-X. (XLSX 13917 kb)
Rights and permissions
About this article
Cite this article
Best, J., Blair, D., Knell, J. et al. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat Immunol 14, 404–412 (2013). https://doi.org/10.1038/ni.2536
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2536
This article is cited by
-
The CUL5 E3 ligase complex negatively regulates central signaling pathways in CD8+ T cells
Nature Communications (2024)
-
Transcription factors in chimeric antigen receptor T-cell development
Human Cell (2024)
-
Targeting PGLYRP1 promotes antitumor immunity while inhibiting autoimmune neuroinflammation
Nature Immunology (2023)
-
Distinct transcriptomic and epigenomic modalities underpin human memory T cell subsets and their activation potential
Communications Biology (2023)
-
Regulation of effector and memory CD8 + T cell differentiation: a focus on orphan nuclear receptor NR4A family, transcription factor, and metabolism
Immunologic Research (2023)