Abstract
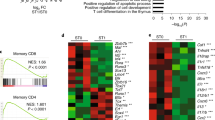
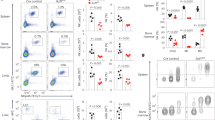
The transcriptional repressor Blimp-1 mediates the terminal differentiation of many cell types, including T cells. Here we identified Hobit (Znf683) as a previously unrecognized homolog of Blimp-1 that was specifically expressed in mouse natural killer T cells (NKT cells). Through studies of Hobit-deficient mice, we found that Hobit was essential for the formation of mature thymic NKT cells. In the periphery, Hobit repressed the accumulation of interferon-γ (IFN-γ)-producing NK1.1lo NKT cells at steady state. After antigenic stimulation, Hobit repressed IFN-γ expression, whereas after innate stimulation, Hobit induced granzyme B expression. Thus, reminiscent of the function of Blimp-1 in other lymphocytes, Hobit controlled the maintenance of quiescent, fully differentiated NKT cells and regulated their immediate effector functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bendelac, A., Savage, P.B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Bendelac, A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182, 2091–2096 (1995).
Mendiratta, S.K. et al. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity 6, 469–477 (1997).
Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Benlagha, K., Kyin, T., Beavis, A., Teyton, L. & Bendelac, A. A thymic precursor to the NK T cell lineage. Science 296, 553–555 (2002).
Pellicci, D.G. et al. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1–CD4+ CD1d-dependent precursor stage. J. Exp. Med. 195, 835–844 (2002).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Savage, A.K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Matsuda, J.L. et al. T-bet concomitantly controls migration, survival, and effector functions during the development of Vα14i NKT cells. Blood 107, 2797–2805 (2006).
Townsend, M.J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 20, 477–494 (2004).
Ohteki, T. et al. The transcription factor interferon regulatory factor 1 (IRF-1) is important during the maturation of natural killer 1.1+ T cell receptor-α/β+ (NK1+ T) cells, natural killer cells, and intestinal intraepithelial T cells. J. Exp. Med. 187, 967–972 (1998).
Kim, P.J. et al. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 177, 6650–6659 (2006).
Mycko, M.P. et al. Selective requirement for c-Myc at an early stage of Vα14i NKT cell development. J. Immunol. 182, 4641–4648 (2009).
Lazarevic, V. et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009).
Walunas, T.L., Wang, B., Wang, C.R. & Leiden, J.M. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J. Immunol. 164, 2857–2860 (2000).
Lacorazza, H.D. et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–449 (2002).
Stanic, A.K. et al. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J. Immunol. 172, 2265–2273 (2004).
Bikoff, E.K., Morgan, M.A. & Robertson, E.J. An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 19, 379–385 (2009).
Nutt, S.L., Fairfax, K.A. & Kallies, A. BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 7, 923–927 (2007).
Kallies, A., Xin, A., Belz, G.T. & Nutt, S.L. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31, 283–295 (2009).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Shin, H. et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009).
Kallies, A. et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood 117, 1869–1879 (2011).
Smith, M.A. et al. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J. Immunol. 185, 6058–6067 (2010).
Hertoghs, K.M. et al. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J. Clin. Invest. 120, 4077–4090 (2010).
Kreslavsky, T. et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA 106, 12453–12458 (2009).
Stenström, M., Skold, M., Andersson, A. & Cardell, S.L. Natural killer T-cell populations in C57BL/6 and NK1.1 congenic BALB.NK mice-a novel thymic subset defined in BALB.NK mice. Immunology 114, 336–345 (2005).
Weinreich, M.A., Odumade, O.A., Jameson, S.C. & Hogquist, K.A. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11, 709–716 (2010).
McNab, F.W. et al. Peripheral NK1.1 NKT cells are mature and functionally distinct from their thymic counterparts. J. Immunol. 179, 6630–6637 (2007).
Crowe, N.Y. et al. Glycolipid antigen drives rapid expansion and sustained cytokine production by NK T cells. J. Immunol. 171, 4020–4027 (2003).
Wingender, G., Krebs, P., Beutler, B. & Kronenberg, M. Antigen-specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178-dependent and is correlated with antigenic potency. J. Immunol. 185, 2721–2729 (2010).
Fehniger, T.A. et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 26, 798–811 (2007).
Bubić, I. et al. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78, 7536–7544 (2004).
Tyznik, A.J. et al. Cutting edge: the mechanism of invariant NKT cell responses to viral danger signals. J. Immunol. 181, 4452–4456 (2008).
Paget, C. et al. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity 27, 597–609 (2007).
Kohlmeier, J.E., Cookenham, T., Roberts, A.D., Miller, S.C. & Woodland, D.L. Type I interferons regulate cytolytic activity of memory CD8+ T cells in the lung airways during respiratory virus challenge. Immunity 33, 96–105 (2010).
Brigl, M., Bry, L., Kent, S.C., Gumperz, J.E. & Brenner, M.B. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 4, 1230–1237 (2003).
Brigl, M. et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J. Exp. Med. 208, 1163–1177 (2011).
Nakamatsu, M. et al. Role of interferon-γ in Vα14+ natural killer T cell-mediated host defense against Streptococcus pneumoniae infection in murine lungs. Microbes Infect. 9, 364–374 (2007).
Cimmino, L. et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol. 181, 2338–2347 (2008).
Acknowledgements
We thank L. Boon (Bioceros) for antibodies; J.C. Zúñiga-Pflücker (Sunnybrook Research Institute) for OP9-DL1 cells; the staff of the animal facility of the Academic Medical Center for animal care; A. de Bruin for technical assistance; B. Hooibrink, E. Mul and F. van Alphen for cell sorting; D. Edo-Matas for help with phylogenetic analysis; and A. Kallies for critical reading of the manuscript. Supported by The Netherlands Organization of Scientific Research (M.A.N. and R.A.W.v.L.) and the Landsteiner Foundation for Blood Transfusion Research (N.A.M.K.).
Author information
Authors and Affiliations
Contributions
K.P.J.M.v.G., N.A.M.K., K.M.L.H. and F.M.W. did experiments; K.P.J.M.v.G., S.J., J.H., M.A.N. and R.A.W.v.L. designed experiments; and K.P.J.M.v.G., M.A.N. and R.A.W.v.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 1347 kb)
Rights and permissions
About this article
Cite this article
van Gisbergen, K., Kragten, N., Hertoghs, K. et al. Mouse Hobit is a homolog of the transcriptional repressor Blimp-1 that regulates NKT cell effector differentiation. Nat Immunol 13, 864–871 (2012). https://doi.org/10.1038/ni.2393
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2393
This article is cited by
-
Cancer-specific tissue-resident memory T-cells express ZNF683 in colorectal cancer
British Journal of Cancer (2023)
-
Alternative splicing of medaka bcl6aa and its repression by Prdm1a and Prdm1b
Fish Physiology and Biochemistry (2021)
-
Tissue-resident memory CD8+ T cells shape local and systemic secondary T cell responses
Nature Immunology (2020)
-
Transcription factor YY1 is essential for iNKT cell development
Cellular & Molecular Immunology (2019)
-
Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation
Nature Immunology (2019)