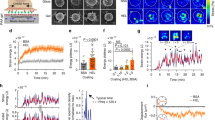
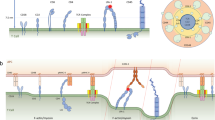
Abstract
Immune synapses form between T cells and antigen-presenting cells (APCs). Increasing evidence suggests synapses must form flexibly to accommodate ongoing motility and displacement of the synapse. Here, time-lapse total internal reflection fluorescence (TIRF) microscopy showed that signaling via the T cell antigen receptor (TCR) occurred during synapse translation. TCR microclusters in motile synapses did not flow directly into supramolecular activating complexes (SMACs) but were directed, independently of myosin II contractility, toward an F-actin-poor 'sink' region. Inward microcluster flow often followed collapse of the leading edge, which suggested that actin depolymerization regulated microcluster flow and the formation of SMACs. The coordination of TCR movement with the translocation of this 'sink' shows how T cells coordinate TCR signaling and microcluster flow in dynamic physiological synapses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Krummel, M.F., Sjaastad, M.D., Wülfing, C. & Davis, M.M. Differential clustering of CD4 and CD3ζ during T cell recognition. Science 289, 1349–1352 (2000).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Wülfing, C. & Davis, M.M.A. Receptor/cytoskeletal movement triggered by costimulation during T cell activation. Science 282, 2266–2269 (1998).
Monks, C.R.F., Freiberg, B.A., Kupfer, H., Sciaky, N. & Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998).
Yokosuka, T. et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 (2005).
Varma, R., Campi, G., Yokosuka, T., Saito, T. & Dustin, M.L. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 (2006).
Mossman, K.D., Campi, G., Groves, J.T. & Dustin, M.L. Altered TCR signaling from geometrically repatterned immunological synapses. Science 310, 1191–1193 (2005).
Kaizuka, Y., Douglass, A.D., Varma, R., Dustin, M.L. & Vale, R.D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl. Acad. Sci. USA 104, 20296–20301 (2007).
Dustin, M.L. & Springer, T.A. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature 341, 619–624 (1989).
Miller, M.J., Safrina, O., Parker, I. & Cahalan, M.D. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J. Exp. Med. 200, 847–856 (2004).
Mempel, T.R., Henrickson, S.E. & von Andrian, U.H. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004).
Celli, S., Garcia, Z. & Bousso, P. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J. Exp. Med. 202, 1271–1278 (2005).
Gunzer, M. et al. Antigen presentation in extracellular matrix: interactions of T cells with dendritic cells are dynamic, shortlived, and sequential. Immunity 13, 323–332 (2000).
Campi, G., Varma, R. & Dustin, M.L. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202, 1031–1036 (2005).
Lee, K.-H. et al. The immunological synapse balances T cell receptor signaling and degradation. Science 302, 1218–1222 (2003).
Friedman, R.S., Beemiller, P., Sorensen, C.M., Jacobelli, J. & Krummel, M.F. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J. Exp. Med. 207, 2733–2749 (2010).
Skokos, D. et al. Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat. Immunol. 8, 835–844 (2007).
Negulescu, P.A., Krasieva, T.B., Khan, A., Kerschbaum, H.H. & Cahalan, M.D. Polarity of T cell shape, motility, and sensitivity to antigen. Immunity 4, 421–430 (1996).
Sims, T.N. et al. Opposing effects of PKCθ and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785 (2007).
Weiss, A., Imboden, J., Shoback, D. & Stobo, J. Role of T3 surface molecules in human T-cell activation: T3-dependent activation results in an increase in cytoplasmic free calcium. Proc. Natl. Acad. Sci. USA 81, 4169–4173 (1984).
Ehrlich, J.S., Hansen, M.D.H. & Nelson, W.J. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev. Cell 3, 259–270 (2002).
Beemiller, P., Jacobelli, J. & Krummel, M.F. Imaging and analysis of OT1 T cell activation on lipid bilayers. Nat. Protoc. published online, doi:10.1038/protex.2012.028 (1 July 2012).
Jacobelli, J., Chmura, S.A., Buxton, D.B., Davis, M.M. & Krummel, M.F. A single class II myosin modulates T cell motility and stopping, but not synapse formation. Nat. Immunol. 5, 531–538 (2004).
Ilani, T., Vasiliver-Shamis, G., Vardhana, S., Bretscher, A. & Dustin, M.L. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat. Immunol. 10, 531–539 (2009).
Yu, Y., Fay, N.C., Smoligovets, A.A., Wu, H.-J. & Groves, J.T. Myosin IIA modulates T cell receptor transport and CasL phosphorylation during early immunological synapse formation. PLoS ONE 7, e30704 (2012).
Yi, J., Wu, X.S., Crites, T. & Hammer, J.A. Actin retrograde flow and acto-myosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T-cells. Mol. Biol. Cell 23, 834–852 (2012).
Straight, A.F. et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743–1747 (2003).
Kolega, J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem. Biophys. Res. Commun. 320, 1020–1025 (2004).
Sakamoto, T., Limouze, J., Combs, C.A., Straight, A.F. & Sellers, J.R. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry 44, 584–588 (2005).
Jacobelli, J., Bennett, F.C., Pandurangi, P., Tooley, A.J. & Krummel, M.F. Myosin-IIA and ICAM-1 regulate the interchange between two distinct modes of T cell migration. J. Immunol. 182, 2041–2050 (2009).
Jacobelli, J. et al. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA–regulated adhesions. Nat. Immunol. 11, 953–961 (2010).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Bubb, M., Senderowicz, A., Sausville, E., Duncan, K. & Korn, E. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 269, 14869–14871 (1994).
Bubb, M., Spector, I., Beyer, B.B. & Fosen, K.M. Effects of Jasplakinolide on the Kinetics of Actin Polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275, 5163–5170 (2000).
Bunnell, S.C., Kapoor, V., Trible, R.P., Zhang, W. & Samelson, L.E. Dynamic actin polymerization drives T cell receptor–Induced spreading: a role for the signal transduction adaptor LAT. Immunity 14, 315–329 (2001).
Vallotton, P., Gupton, S.L., Waterman-Storer, C.M. & Danuser, G. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA 101, 9660–9665 (2004).
Delorme, V. et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell 13, 646–662 (2007).
Eibert, S.M. et al. Cofilin peptide homologs interfere with immunological synapse formation and T cell activation. Proc. Natl. Acad. Sci. USA 101, 1957–1962 (2004).
Hogquist, K.A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Friedman, R.S., Jacobelli, J. & Krummel, M.F. Surface-bound chemokines capture and prime T cells for synapse formation. Nat. Immunol. 7, 1101–1108 (2006).
Rogers, S.S., Waigh, T.A., Zhao, X. & Lu, J.R. Precise particle tracking against a complicated background: polynomial fitting with Gaussian weight. Phys. Biol. 4, 220–227 (2007).
Grynkiewicz, G., Poenie, M. & Tsien, R. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 (1985).
Acknowledgements
We thank S. Rogers (University of Manchester) for the polynomial fit Gaussian weight function; R. Wedlich-Soldner (Max Planck Institute of Biochemistry) for Lifeact-GFP; B. Lillemeier (Salk Institute) and M. Davis (Stanford University) for the histidine-tagged ICAM construct; M. Werner and K. Austgen for assistance in preparing histidine-tagged ICAM; and J. Altman (US National Institutes of Health Tetramer Facility at Emory University) for biotinylated pMHC monomers. Supported by the Cancer Research Institute (P.B.) and the US National Institutes of Health (AI52116 to M.F.K.).
Author information
Authors and Affiliations
Contributions
P.B. and M.F.K. designed the experiments for Figures 1,2,3 and 5,6,7 and P.B. did these experiments; P.B., J.J. and M.F.K. designed the experiments for Figure 4 and P.B. and J.J. did these experiments; P.B. wrote the manuscript; J.J. contributed to editing of the manuscript; and M.F.K. edited and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 759 kb)
Supplementary Movie 1
Microcluster flow couples with cell motility. (MP4 1326 kb)
Supplementary Movie 2
cSMAC formation is independent of synapse motility. (MP4 2430 kb)
Supplementary Movie 3
Myosin II inhibition does not impair motile synapse formation under non-detrimental experimental conditions. (MP4 2975 kb)
Supplementary Movie 4
Synapse formation by control, blebbistatin treated and conditional myosin II knockout cells. (MP4 2500 kb)
Supplementary Movie 5
Cell footprint contraction coordinated TCR microcluster flows. (MP4 3098 kb)
Supplementary Movie 6
TCR microclusters flow into zones of actin depolymerization. (MP4 222 kb)
Supplementary Movie 7
TCR microclusters and cSMAC movement are directed to an F-actin-poor sink region in motile synapses. (MP4 3057 kb)
Rights and permissions
About this article
Cite this article
Beemiller, P., Jacobelli, J. & Krummel, M. Integration of the movement of signaling microclusters with cellular motility in immunological synapses. Nat Immunol 13, 787–795 (2012). https://doi.org/10.1038/ni.2364
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2364
This article is cited by
-
In vitro tracking and intracellular protein distribution in immunology
Immunology & Cell Biology (2017)
-
TIRF imaging of Fc gamma receptor microclusters dynamics and signaling on macrophages during frustrated phagocytosis
BMC Immunology (2016)
-
Actin polymerization‐dependent activation of Cas‐L promotes immunological synapse stability
Immunology & Cell Biology (2016)
-
Origins of the cytolytic synapse
Nature Reviews Immunology (2016)
-
The bullseye synapse formed between CD4+ T‐cell and staphylococcal enterotoxin B‐pulsed dendritic cell is a suppressive synapse in T‐cell response
Immunology & Cell Biology (2015)