Abstract
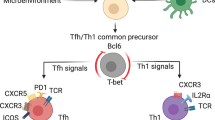
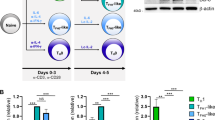
The transcription factors T-bet and Bcl-6 are required for the establishment of a T helper type 1 cell (TH1 cell) and follicular helper T cell (TFH cell) gene-expression profile, respectively. Here we found that high concentrations of interleukin 2 (IL-2) inhibited Bcl-6 expression in polarized TH1 cells. Mechanistically, the low concentrations of Bcl-6 normally found in effector TH1 cells did not repress its target genes because a T-bet–Bcl-6 complex masked the Bcl-6 DNA-binding domain. TH1 cells increased their Bcl-6/T-bet ratio in response to limiting IL-2 conditions, which allowed excess Bcl-6 to repress its direct target Prdm1 (which encodes the transcriptional repressor Blimp-1). The Bcl-6-dependent repression of Blimp-1 effectively induced a partial TFH profile because Blimp-1 directly repressed a subset of TFH signature genes, including Cxcr5. Thus, IL-2-signaling regulates the Bcl-6–Blimp-1 axis in TH1 cells to maintain flexibility with a TFH cell–like gene profile.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Murphy, K.M. & Stockinger, B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat. Immunol. 11, 674–680 (2010).
O'Shea, J.J. & Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010).
Reinhardt, R.L., Kang, S.J., Liang, H.E. & Locksley, R.M. T helper cell effector fates–who, how and where? Curr. Opin. Immunol. 18, 271–277 (2006).
Zhou, L., Chong, M.M. & Littman, D.R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009).
Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Peng, S.L. The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol. Immunol. 3, 87–95 (2006).
Lazarevic, V. & Glimcher, L.H. T-bet in disease. Nat. Immunol. 12, 597–606 (2011).
Ivanov, I.I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Johnston, R.J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009).
Nurieva, R.I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009).
Szabo, S.J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000).
Yu, D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009).
Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Beima, K.M. et al. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J. Biol. Chem. 281, 11992–12000 (2006).
Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Lewis, M.D., Miller, S.A., Miazgowicz, M.M., Beima, K.M. & Weinmann, A.S. T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol. Cell. Biol. 27, 8510–8521 (2007).
Miller, S.A., Huang, A.C., Miazgowicz, M.M., Brassil, M.M. & Weinmann, A.S. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 22, 2980–2993 (2008).
Miller, S.A., Mohn, S.E. & Weinmann, A.S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 (2010).
Szabo, S.J. et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science 295, 338–342 (2002).
Hwang, E.S., Szabo, S.J., Schwartzberg, P.L. & Glimcher, L.H. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005).
Lazarevic, V. et al. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12, 96–104 (2011).
Mehta, D.S., Wurster, A.L., Weinmann, A.S. & Grusby, M.J. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc. Natl. Acad. Sci. USA 102, 2016–2021 (2005).
Oestreich, K.J., Huang, A.C. & Weinmann, A.S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 208, 1001–1013 (2011).
Basso, K. & Dalla-Favera, R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 105, 193–210 (2010).
Mascle, X., Albagli, O. & Lemercier, C. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 300, 391–396 (2003).
Vinuesa, C.G., Tangye, S.G., Moser, B. & Mackay, C.R. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 5, 853–865 (2005).
Pipkin, M.E. et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90 (2010).
Choi, Y.S. et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011).
Liao, W., Lin, J.X. & Leonard, W.J. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 23, 598–604 (2011).
Merkenschlager, M. & von Boehmer, H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J. Exp. Med. 207, 1347–1350 (2010).
Stittrich, A.B. et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 11, 1057–1062 (2011).
Mandal, M. et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12, 1212–1220 (2011).
Yang, X.P. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 (2011).
Riou, C. et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J. Exp. Med. 204, 79–91 (2007).
Hurtz, C. et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med. 208, 2163–2174 (2011).
Crotty, S., Johnston, R.J. & Schoenberger, S.P. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11, 114–120 (2010).
Martins, G. & Calame, K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26, 133–169 (2008).
Hegazy, A.N. et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 32, 116–128 (2010).
Koch, M.A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009).
Wang, Y., Su, M.A. & Wan, Y.Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348 (2011).
Liao, W., Lin, J.X., Wang, L., Li, P. & Leonard, W.J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12, 551–559 (2011).
Pepper, M., Pagan, A.J., Igyarto, B.Z., Taylor, J.J. & Jenkins, M.K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Bauquet, A.T. et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10, 167–175 (2009).
Ise, W. et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12, 536–543 (2011).
Lee, S.K. et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J. Exp. Med. 208, 1377–1388 (2011).
Acknowledgements
We thank the National Cancer Institute preclinical repository for IL-2 and anti-IL-4; members of the Weinmann laboratory for discussions; and M. Wijaranakula for technical assistance. Supported by the National Institute of Allergy and Infectious Diseases (AI061061 and AI07272 to A.S.W.) and the American Cancer Society (RSG-09-045-01-DDC to A.S.W.).
Author information
Authors and Affiliations
Contributions
K.J.O. and A.S.W. designed and did experiments, analyzed data and wrote the manuscript; and S.E.M contributed to the experiments in Figures 1c and 2b,e, and Supplementary Figure 4a,b.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Table 1 (PDF 2220 kb)
Rights and permissions
About this article
Cite this article
Oestreich, K., Mohn, S. & Weinmann, A. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 13, 405–411 (2012). https://doi.org/10.1038/ni.2242
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2242
This article is cited by
-
IL6 suppresses vaccine responses in neonates by enhancing IL2 activity on T follicular helper cells
npj Vaccines (2023)
-
CD4+ T cell memory
Nature Immunology (2023)
-
Aiolos represses CD4+ T cell cytotoxic programming via reciprocal regulation of TFH transcription factors and IL-2 sensitivity
Nature Communications (2023)
-
CTLA4 protects against maladaptive cytotoxicity during the differentiation of effector and follicular CD4+ T cells
Cellular & Molecular Immunology (2023)
-
TCF-1: a maverick in T cell development and function
Nature Immunology (2022)