Abstract
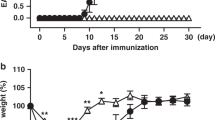
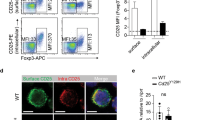
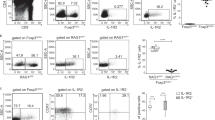
Mice deficient in the adaptor Ndfip1 develop inflammation at sites of environmental antigen exposure. We show here that such mice had fewer inducible regulatory T cells (iTreg cells). In vitro, Ndfip1-deficient T cells expressed normal amounts of the transcription factor Foxp3 during the first 48 h of iTreg cell differentiation; however, this expression was not sustained. Abortive Foxp3 expression was caused by production of interleukin 4 (IL-4) by Ndfip1−/− cells. We found that Ndfip1 expression was transiently upregulated during iTreg cell differentiation in a manner dependent on transforming growth factor-β (TGF-β). Once expressed, Ndfip1 promoted degradation of the transcription factor JunB mediated by the E3 ubiquitin ligase Itch, thus preventing IL-4 production. On the basis of our data, we propose that TGF-β signaling induces Ndfip1 expression to silence IL-4 production, thus permitting iTreg cell differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Bluestone, J.A. & Abbas, A.K. Natural versus adaptive regulatory T cells. Nat. Rev. Immunol. 3, 253–257 (2003).
Curotto de Lafaille, M.A. & Lafaille, J.J. Natural and adaptive Foxp3+ regulatory T cells: more of the same or division of labor. Immunity 30, 626–635 (2009).
Brunkow, M.E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001).
Bennett, C.L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001).
Wildin, R.S. et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27, 18–20 (2001).
Fontenot, J.D., Gavin, M.A. & Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Khattri, R., Cox, T., Yasayko, S.A. & Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4, 337–342 (2003).
Josefowicz, S.Z. & Rudensky, A.Y. Control of regulatory T cell lineage commitment and maintenance. Immunity 30, 616–625 (2009).
Malek, T.R. & Castro, I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33, 153–165 (2010).
Xu, L., Kitani, A. & Strober, W. Molecular mechanisms regulating TGF-beta-induced Foxp3 expression. Mucosal Immunol. 3, 230–238 (2010).
Sakaguchi, S., Wing, K. & Miyara, M. Regulatory T cells—a brief history and perpective. Eur. J. Immunol. 37, S116–S123 (2007).
Chen, W. et al. Conversion of peripheral CD4+CD25 naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Kretschmer, K. et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6, 1219–1227 (2005).
Apostolou, I. & von Boehmer, H. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 199, 1401–1408 (2004).
Fantini, M.C. et al. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25 T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 172, 5149–5153 (2004).
Park, H.B., Paik, D.J., Jang, E., Hong, S. & Youn, J. Acquisition of anergic and suppressive activities in transforming growth factor-beta-costimulated CD4+CD25 T cells. Int. Immunol. 16, 1203 (2004).
Wan, Y.Y. & Flavell, R.A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. USA 102, 5126–5131 (2005).
Dardalhon, V. et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells, and together with TGF-β generates IL-9+ IL-10+ Foxp3− effector T cells. Nat. Immunol. 9, 1347–1355 (2008).
Gorelik, L., Fields, P.E. & Flavell, R.A. TGF-β inhibits Th type 2 development through inhibition of GATA-3 expression. J. Immunol. 165, 4773–4777 (2000).
Tone, Y. et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202 (2007).
Hefferan, T.E. et al. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J. Biol. Chem. 275, 20255–20259 (2000).
Venuprasad, K. et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol. 9, 245–253 (2008).
Cao, Z. et al. Kruppel-like factor KLF10 targets transforming growth factor-β to regulate CD4+CD25 T cells and T regulatory cells. J. Biol. Chem. 11, 24914–24924 (2009).
Oliver, P.M. et al. Ndfip1 protein promotes the function of Itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940 (2006).
Rooney, J.W., Hoey, T. & Glimcher, L.H. Coordinate and cooperative roles for NF-AT and AP-1 in the regulation of murine IL-4 gene. Immunity 2, 473–483 (1995).
Li-Weber, M., Giasi, M. & Krammer, P.H. Involvement of Jun and Rel proteins in up-regulation of interleukin-4 gene activity by the T cell accessory molecule CD28. J. Biol. Chem. 273, 32460–32466 (1998).
Fang, D. et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3, 281–287 (2002).
Ramon, H.E. et al. The ubiquitin ligase adaptor Ndfip1 regulates T cell-mediated gastrointestinal inflammation and inflammatory bowel disease susceptibility. Mucosal Immunol. 4, 314–324 (2011).
Liu, Y. et al. A critical function for TGF-β signaling in the development of natural CD4+ CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9, 632–640 (2008).
Fontenot, J.D., Dooley, J.L., Farr, A.G. & Rudensky, A.Y. Developmental regulation of Foxp3 expression during ontogeny. J. Exp. Med. 202, 901–906 (2005).
Coombes, J.L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Sun, C.M. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204, 1775–1785 (2007).
Siddiqui, K.R. & Powrie, F. CD103+ GALT DCs promote Foxp3+ regulatory T cells. Mucosal Immunol. 1 (suppl. 1), S34–S38 (2008).
Thornton, A.M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010).
Verhagen, J. & Wraith, D.C. Comment on “Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells”. J. Immunol. 185, 7129 (2010).
Thornton, A.M. & Shevach, E.M. Response to comment on “Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells”. J. Immunol. 185, 7130 (2010).
Wei, J. et al. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. USA 104, 18169–18174 (2007).
Mantel, P.Y. et al. GATA3-driven Th2 responses inhibit TGF-β1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 5, e329 (2007).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Selvaraj, R.K. & Geiger, T.L. A kinetic and dynamic analysis of Foxp3 induced in T cells by TGF-β. J. Immunol. 178, 7667–7677 (2007).
Chen, Q., Kim, Y.C., Laurence, A., Punkosdy, G.A. & Shevach, E.M. IL-2 controls the stability of Foxp3 expression in TGF-β-induced Foxp3+ T cells in vivo. J. Immunol. 186, 6329–6337 (2011).
Liao, W. et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 9, 1288–1296 (2008).
Jonk, L.J., Itoh, S., Heldin, C.H., ten Dijke, P. & Kruijer, W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152 (1998).
Liang, Q. et al. IL-2 and IL-4 stimulate MEK1 expression and contribute to T cell resistance against suppression by TGF-β and IL-10 in asthma. J. Immunol. 185, 5704–5713 (2010).
Wang, J., Ioan-Facsinay, A., van der Voort, E.I., Huizinga, T.W. & Toes, R.E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 37, 129–138 (2007).
Ramsdell, F., Jenkins, M., Dinh, Q. & Fowlkes, B.J. The majority of CD4+CD8thymocytes are functionally immature. J. Immunol. 147, 1779–1785 (1991).
Cote-Sierra, J. et al. Interleukin-2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 101, 3880–3885 (2004).
Li, B., Tournier, C., Davis, R.J. & Flavell, R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18, 420–432 (1999).
Park, J., Kim, S.H., Li, Q., Chang, Y.T. & Kim, T.S. Inhibition of interleukin-4 production in activated T cells via the downregulation of AP-1/NF-AT activation by N-lauroyl-D-erythro-sphingosine and N-lauroyl-d-erythro-C20-sphingosine. Biochem. Pharmacol. 71, 1229–1239 (2006).
Acknowledgements
We thank H. Ramon for discussions and for help with setting up iTreg cell culture conditions; A. Bhandoola for critical reading of the manuscript; J.K. Burkhardt for discussions; A. Laroche for technical assistance; and the staff of the flow cytometry core at the University of Pennsylvania. Supported by the U.S. National Institutes of Health (RO3 AR057144, 1F32AI085837 and R01AI093566).
Author information
Authors and Affiliations
Contributions
A.M.B. designed and did experiments and wrote the manuscript; N.R.-H., C.R.R. and E.A.N. did experiments and contributed data; and P.M.O. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Methods (PDF 1038 kb)
Rights and permissions
About this article
Cite this article
Beal, A., Ramos-Hernández, N., Riling, C. et al. TGF-β induces the expression of the adaptor Ndfip1 to silence IL-4 production during iTreg cell differentiation. Nat Immunol 13, 77–85 (2012). https://doi.org/10.1038/ni.2154
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2154
This article is cited by
-
Ndfip1 restricts mTORC1 signalling and glycolysis in regulatory T cells to prevent autoinflammatory disease
Nature Communications (2017)
-
IL-1β induced HIF-1α inhibits the differentiation of human FOXP3+ T cells
Scientific Reports (2017)
-
The transcription factor musculin promotes the unidirectional development of peripheral Treg cells by suppressing the TH2 transcriptional program
Nature Immunology (2017)
-
Ndfip1 restricts Th17 cell potency by limiting lineage stability and proinflammatory cytokine production
Scientific Reports (2017)
-
Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination
Nature Immunology (2016)