Abstract
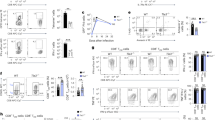
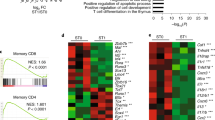
It is established that the transcription factor E2A and its antagonist Id3 modulate the checkpoints consisting of the precursor to the T cell antigen receptor (pre-TCR) and the TCR. Here we demonstrate that Id3 expression was higher beyond the pre-TCR checkpoint, remained high in naive T cells and showed a bimodal pattern in the effector-memory population. We show how E2A promoted T lineage specification and how pre-TCR-mediated signaling affected E2A genome-wide occupancy. Thymi in Id3-deficient mice had aberrant development of effector-memory cells, higher expression of the chemokine receptor CXCR5 and the transcriptional repressor Bcl-6 and, unexpectedly, T cell–B cell conjugates and B cell follicles. Collectively, our data show how E2A acted globally to orchestrate development into the T lineage and that Id3 antagonized E2A activity beyond the pre-TCR checkpoint to enforce the naive fate of T cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
Change history
25 June 2012
In the version of this article initially published, the GEO accession code for the ChIP–Seq data set was not included. The code is GSE30518. The error has been corrected in the HTML and PDF versions of the article.
References
Kreslavsky, T., Gleimer, M., Garbe, A.I. & von Boehmer, H. αβ versus γδ fate choice: counting the T-cell lineage at the branch point. Immunol. Rev. 238, 169–181 (2010).
Allman, D. et al. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4, 168–174 (2003).
Taghon, T., Yui, M.A., Pant, R., Diamond, R.A. & Rothenberg, E.V. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006).
Yashiro-Ohtani, Y. et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 23, 1665–1676 (2009).
Carpenter, A.C. & Bosselut, R. Decision checkpoints in the thymus. Nat. Immunol. 8, 666–673 (2010).
Murre, C. Developmental trajectories in early hematopoiesis. Genes Dev. 15, 2366–2370 (2009).
Zhang, J., Kalkum, M., Yamamura, S., Chait, B.T. & Roeder, R.G. E-protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305, 1286–1289 (2004).
Massari, M.E. & Murre, C. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 18, 3130–3139 (2000).
Beck, K., Peak, M.M., Ota, T., Nemazee, D. & Murre, C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 206, 2271–2284 (2009).
Benezra, R., Davis, R.L., Lockshon, D., Turner, D.L. & Weintraub, H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61, 49–59 (1990).
Kee, B.L. E and Id proteins branch out. Nat. Rev. Immunol. 9, 175–184 (2009).
Barndt, R.J., Dai, M. & Zhuang, Y. Functions of E2A/HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20, 6677–6685 (2000).
Ikawa, T., Kawamoto, H., Goldrath, A.W. & Murre, C. E proteins and Notch signalling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 203, 1329–1342 (2006).
Herblot, S., Steff, A.M., Hugo, P., Aplan, P.D. & Hoang, T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Tα chain expression. Nat. Immunol. 1, 138–144 (2000).
Agata, Y. et al. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 27, 871–884 (2007).
Schwartz, R. et al. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediate signalling and T-lineage development. Proc. Natl. Acad. Sci. USA 103, 9976–9981 (2006).
Engel, I., Johns, C., Bain, G., Rivera, R.R. & Murre, C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194, 733–745 (2001).
Bain, G. et al. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2, 165–171 (2001).
Bain, G. et al. Thymocyte differentiation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 190, 1605–1616 (1999).
Jones, M.E. & Zhuang, Y. Acquisition of a functional TCR during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 6, 860–868 (2007).
Rivera, R.R., Johns, C.P., Quan, J., Johnson, R.S. & Murre, C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12, 17–26 (2000).
D'Cruz, L.M., Knell, J., Fujimoto, J.K. & Goldrath, A.W. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11, 240–249 (2010).
Ueda-Hayakawa, I., Mahlios, J. & Zhuang, Y. Id3 restricts the developmental potential of γδ lineage during thymopoiesis. J. Immunol. 182, 5306–5316 (2007).
Alonzo, E.S. et al. Development of promyelocytic zinc finger and ThPOK-expressing innate γδ T cells is controlled by strength of TCR signaling and Id3. J. Immunol. 184, 1268–1279 (2010).
Lee, S.Y., Stadanlick, J., Kappes, D.J. & Wiest, D.L. Towards a molecular understanding of the differential signals regulating αβ/γδ T lineage choice. Semin. Immunol. 4, 237–246 (2010).
Verykokakis, M. et al. Inhibitor of DNA binding 3 limits development of murine slam-associated adpator protein-dependent “innate” γδ T cells. PLoS ONE 15, e9303 (2010).
Verykokakis, M., Boos, M.D., Bendelac, A. & Kee, B.L. SAP protein-dependent natural killer T-like cells regulate the development of CD8+ T cells with innate lymphocyte characteristics. Immunity 27, 203–215 (2010).
Maruyama, T. et al. Control of the differentiation of regulatory T cells and TH17 cells by the DNA-binding inhibitor Id3. Nat. Immunol. 12, 86–95 (2011).
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Lin, Y.C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 28, 576–589 (2010).
Wadman, I.A. et al. The LIM-only protein LMO2 is a bridging molecule assembling an erythroid, DNA binding complex which includes the TAL1, E47, GATA-1 and Ldb1/Nl1 proteins. EMBO J. 16, 3145–3157 (1997).
Palacios, E.H. & Weiss, A. Distinct roles for Syk and ZAP-70 during early thymocyte development. J. Exp. Med. 204, 1703–1715 (2007).
Cannons, J.L. et al. Optimal germinal center responses require multistage T cell: B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity 32, 253–265 (2010).
Johnston, R.J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2, 1006–1010 (2009).
Nurieva, R.I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009).
Wolfer, A. et al. Inactivation of Notch1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2, 235–241 (2001).
Weinreich, M.A., Odumade, O.A., Jameson, S.C. & Hogquist, K.A. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11, 709–716 (2010).
Purton, J.F. et al. Antiviral CD4+ memory T cells are IL-15 dependent. J. Exp. Med. 204, 951–961 (2007).
Georgescu, C. et al. A gene regulatory network armature for T lymphocyte specification. Proc. Natl. Acad. Sci. USA 105, 20100–20105 (2008).
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte differentiation. Cell 111, 621–633 (2002).
Eyquem, S., Chemin, K., Fasseu, M. & Bories, J.C. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor β locus. Proc. Natl. Acad. Sci. USA 44, 15712–15717 (2004).
Sakaguchi, N. et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426, 454–460 (2003).
Li, H., Dai, M. & Zhuang, Y.A. T cell intrinsic role of Id3 in a mouse model for primary Sjögren's syndrome. Immunity 4, 551–560 (2004).
Saini, M. et al. Regulation of Zap70 expression during thymocyte development enables temporal separation of CD4 and CD8 repertoire selection at different signaling thresholds. Sci. Signal. 3, 114–121 (2010).
Quong, M.W. et al. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J. Exp. Med. 199, 1101–1112 (2004).
Acknowledgements
We thank A. Goldrath and members of the Murre laboratory for critical reading of the manuscript; G. Hardiman, M. Harabaglia, J. Sprague and C. Ludka for help with Solexa DNA sequencing; N. Varki and the University of California, San Diego Histology Core for histology; and Y. Zhuang (Duke University) for E2a/GFP and E2af/f mice. Supported by the American Recovery and Investment Act (27775A to Y.C.L.) and the US National Institutes of Health (C.M.).
Author information
Authors and Affiliations
Contributions
M.M. designed and did experiments, analyzed data and wrote portions of the manuscript; R.R.R. generated Id3-GFP mice; K.M. did ChIP analyses; Y.C.L. contributed to the analysis of data from ChIP followed by deep sequencing; Y.A. provided advice on ChIP experiments with small number of cells; and C.M. designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 3166 kb)
Rights and permissions
About this article
Cite this article
Miyazaki, M., Rivera, R., Miyazaki, K. et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol 12, 992–1001 (2011). https://doi.org/10.1038/ni.2086
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2086
This article is cited by
-
How transcription factors drive choice of the T cell fate
Nature Reviews Immunology (2021)
-
Single-cell RNA-sequencing identifies the developmental trajectory of C-Myc-dependent NK1.1− T-bet+ intraepithelial lymphocyte precursors
Mucosal Immunology (2020)
-
Early precursor T cells establish and propagate T cell exhaustion in chronic infection
Nature Immunology (2020)
-
Mucosal T follicular helper cells in SIV-infected rhesus macaques: contributing role of IL-27
Mucosal Immunology (2019)
-
Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia
Leukemia (2018)