Abstract
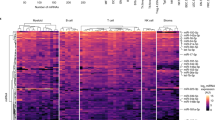
MicroRNAs are small noncoding RNAs that regulate gene expression post-transcriptionally. Here we applied microRNA profiling to 17 human lymphocyte subsets to identify microRNA signatures that were distinct among various subsets and different from those of mouse lymphocytes. One of the signature microRNAs of naive CD4+ T cells, miR-125b, regulated the expression of genes encoding molecules involved in T cell differentiation, including IFNG, IL2RB, IL10RA and PRDM1. The expression of synthetic miR-125b and lentiviral vectors encoding the precursor to miR-125b in naive lymphocytes inhibited differentiation to effector cells. Our data provide an 'atlas' of microRNA expression in human lymphocytes, define subset-specific signatures and their target genes and indicate that the naive state of T cells is enforced by microRNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103, 2257–2261 (2006).
Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 (2005).
O'Connell, R.M., Taganov, K.D., Boldin, M.P., Cheng, G. & Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104, 1604–1609 (2007).
Kaech, S.M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8+ T cell differentiation. Cell 111, 837–851 (2002).
O'Shea, J.J. & Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010).
Lodish, H.F., Zhou, B., Liu, G. & Chen, C.Z. Micromanagement of the immune system by microRNAs. Natl. Rev. 8, 120–130 (2008).
O'Connell, R.M., Rao, D.S., Chaudhuri, A.A. & Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122 (2010).
O'Carroll, D. et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004 (2007).
Cobb, B.S. et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201, 1367–1373 (2005).
Koralov, S.B. et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874 (2008).
Li, Q.J. et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 (2007).
Xiao, C. et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159 (2007).
Rodriguez, A. et al. Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 (2007).
Thai, T.H. et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608 (2007).
O'Connell, R.M. et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 (2010).
Malumbres, R. et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113, 3754–3764 (2009).
O'Connell, R.M. et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA 107, 14235–14240 (2010).
Bousquet, M., Harris, M.H., Zhou, B. & Lodish, H.F. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. USA 107, 21558–21563 (2010).
Ooi, A.G. et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA 107, 21505–21510 (2010).
Gefen, N. et al. HSA-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 24, 89–96 (2010).
Bousquet, M. et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 205, 2499–2506 (2008).
Kuchen, S. et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839 (2010).
Basso, K. et al. Identification of the human mature B cell miRNome. Immunity 30, 744–752 (2009).
Landgraf, P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007).
Cosmi, L. et al. CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur. J. Immunol. 30, 2972–2979 (2000).
Acosta-Rodriguez, E.V. et al. Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat. Immunol. 8, 639–646 (2007).
Banham, A.H. Cell-surface IL-7 receptor expression facilitates the purification of FOXP3+ regulatory T cells. Trends Immunol. 27, 541–544 (2006).
Sallusto, F., Geginat, J. & Lanzavecchia, A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763 (2004).
Kleinschek, M.A. et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 206, 525–534 (2009).
Cobb, B.S. et al. A role for Dicer in immune regulation. J. Exp. Med. 203, 2519–2527 (2006).
Lu, L.F. et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929 (2010).
Li, S.C. et al. Identification of homologous microRNAs in 56 animal genomes. Genomics 96, 1–9 (2010).
Mestdagh, P. et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 36, e143 (2008).
Thomas, M., Lieberman, J. & Lal, A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17, 1169–1174 (2010).
Tan, J.T. et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl. Acad. Sci. USA 98, 8732–8737 (2001).
Rivino, L. et al. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J. Exp. Med. 200, 725–735 (2004).
Purton, J.F. et al. Antiviral CD4+ memory T cells are IL-15 dependent. J. Exp. Med. 204, 951–961 (2007).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 6, 654–659 (2007).
Acknowledgements
We thank P. Dellabona, P. De Candia and S. Monticelli for discussions. Supported by the National Research Fund of Luxembourg (PDR-08-040 to L.W.) and the Fondazione Cariplo.
Author information
Authors and Affiliations
Contributions
R.L.R., G.R. and L.W. designed and did experiments, analyzed data and wrote the manuscript; M.M. and M.C.C. did flow cytometry; R.J.P.B. did bioinformatic analyses; M.C. did statistical analyses; R.S.B., S.C., S.M., P.G., F.M., D.M., V.P. and A.R. did experiments and analyzed data; E.T. provided advice; R.D.F. discussed results, provided advice and commented on the manuscript; J.G. designed and supervised research and wrote the manuscript; and S.A. and M.P. designed the study, supervised research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–4 and Supplementary Methods (PDF 1048 kb)
Supplementary Spreadsheet
RTqPCR raw CT data for all samples profiled. (TXT 218 kb)
Rights and permissions
About this article
Cite this article
Rossi, R., Rossetti, G., Wenandy, L. et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol 12, 796–803 (2011). https://doi.org/10.1038/ni.2057
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2057
This article is cited by
-
Upregulated microRNA-125b-5p in patients with asthma-COPD overlap mediates oxidative stress and late apoptosis via targeting IL6R/TRIAP1 signaling
Respiratory Research (2024)
-
Death Induced by Survival gene Elimination (DISE) correlates with neurotoxicity in Alzheimer’s disease and aging
Nature Communications (2024)
-
The interaction of mast cells with membranes from lung cancer cells induces the release of extracellular vesicles with a unique miRNA signature
Scientific Reports (2023)
-
MicroRNA profile of circulating CD4+ T cells in aged patients with atherosclerosis obliterans
BMC Cardiovascular Disorders (2022)
-
Differential Leukocyte MicroRNA Responses Following Pan T Cell, Allorecognition and Allosecretome-Based Therapeutic Activation
Archivum Immunologiae et Therapiae Experimentalis (2021)