Abstract
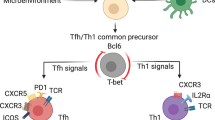
Helper T cells control host defense against pathogens. The receptors for interleukin 12 (IL-12), IL-4 and IL-6 are required for differentiation into the TH1, TH2 and TH17 subsets of helper T cells, respectively. IL-2 signaling via the transcription factor STAT5 controls TH2 differentiation by regulating both the TH2 cytokine gene cluster and expression of Il4ra, the gene encoding the IL-4 receptor α-chain. Here we show that IL-2 regulated TH1 differentiation, inducing STAT5-dependent expression of the IL-12 receptor β2-chain (IL-12Rβ2) and the transcription factor T-bet, with impaired human TH1 differentiation when IL-2 was blocked. TH1 differentiation was also impaired in mouse Il2−/− T cells but was restored by IL-12Rβ2 expression. Consistent with the inhibition of TH17 differentiation by IL-2, treatment with IL-2 resulted in lower expression of the genes encoding the IL-6 receptor α-chain (Il6ra) and the IL-6 signal transducer gp130 (encoded by Il6st), and retroviral transduction of Il6st augmented TH17 differentiation even when IL-2 was present. Thus, IL-2 influences helper T cell differentiation by modulating the expression of cytokine receptors to help specify and maintain differentiated states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
Change history
20 May 2011
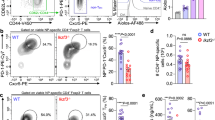
In the version of this article initially published online, the labels along the horizontal axes of the graphs at right in Figure 8b,c were incorrect, and the legend for Figure 8a–c did not correctly specify the times of stimulation. The correct labels are '–' (instead of Il2+/+) and '+' (instead of Il2–/–), and the corrected legend states that the cells were "left unstimulated (–) or stimulated (+) for 4 h (a) or 24 h (b,c) with IL-2." The error has been corrected for the PDF and HTML versions of this article.
References
Zhu, J. & Paul, W.E. CD4 T cells: fates, functions, and faults. Blood 112, 1557–1569 (2008).
Maldonado, R.A. et al. Control of T helper cell differentiation through cytokine receptor inclusion in the immunological synapse. J. Exp. Med. 206, 877–892 (2009).
Szabo, S.J., Dighe, A.S., Gubler, U. & Murphy, K.M. Regulation of the interleukin (IL)-12Rβ2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 185, 817–824 (1997).
Liao, W. et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 9, 1288–1296 (2008).
Nishihara, M. et al. IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int. Immunol. 19, 695–702 (2007).
Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V.K. IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009).
Szabo, S.J., Sullivan, B.M., Peng, S.L. & Glimcher, L.H. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 21, 713–758 (2003).
Ansel, K.M., Djuretic, I., Tanasa, B. & Rao, A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24, 607–656 (2006).
Lin, J.X. & Leonard, W.J. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 19, 2566–2576 (2000).
Leonard, W.J. Cytokines and immunodeficiency diseases. Nat. Rev. Immunol. 1, 200–208 (2001).
Kim, H.P., Imbert, J. & Leonard, W.J. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 17, 349–366 (2006).
Cote-Sierra, J. et al. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 101, 3880–3885 (2004).
Bream, J.H. et al. A distal region in the interferon-γ gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J. Biol. Chem. 279, 41249–41257 (2004).
Reem, G.H. & Yeh, N.H. Interleukin 2 regulates expression of its receptor and synthesis of γ interferon by human T lymphocytes. Science 225, 429–430 (1984).
Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007).
Mullen, A.C. et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science 292, 1907–1910 (2001).
Afkarian, M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 3, 549–557 (2002).
Hwang, E.S., Szabo, S.J., Schwartzberg, P.L. & Glimcher, L.H. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005).
Bird, J.J. et al. Helper T cell differentiation is controlled by the cell cycle. Immunity 9, 229–237 (1998).
Lighvani, A.A. et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc. Natl. Acad. Sci. USA 98, 15137–15142 (2001).
Chang, J.T., Segal, B.M. & Shevach, E.M. Role of costimulation in the induction of the IL-12/IL-12 receptor pathway and the development of autoimmunity. J. Immunol. 164, 100–106 (2000).
Leonard, W.J. et al. A monoclonal antibody that appears to recognize the receptor for human T-cell growth factor; partial characterization of the receptor. Nature 300, 267–269 (1982).
Tsudo, M., Kitamura, F. & Miyasaka, M. Characterization of the interleukin 2 receptor β chain using three distinct monoclonal antibodies. Proc. Natl. Acad. Sci. USA 86, 1982–1986 (1989).
Onishi, M. et al. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18, 3871–3879 (1998).
Yang, Y., Ochando, J.C., Bromberg, J.S. & Ding, Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNγ/Stat1 signals. Blood 110, 2494–2500 (2007).
Kano, S. et al. The contribution of transcription factor IRF1 to the interferon-γ–interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol. 9, 34–41 (2008).
Lee, Y.K. et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009).
Leonard, W.J. & O'Shea, J.J. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16, 293–322 (1998).
Shi, M., Lin, T.H., Appell, K.C. & Berg, L.J. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity 28, 763–773 (2008).
Lazarevic, V. et al. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12, 96–104 (2011).
Imada, K. et al. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 188, 2067–2074 (1998).
Malek, T.R. The biology of interleukin-2. Annu. Rev. Immunol. 26, 453–479 (2008).
Xue, H.H. et al. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. Proc. Natl. Acad. Sci. USA 99, 13759–13764 (2002).
Ben-Sasson, S.Z. et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 106, 7119–7124 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Acknowledgements
We thank G. Robinson and L. Hennighausen (National Institute of Diabetes and Digestive and Kidney Diseases of the US National Institutes of Health) for mice with loxP-flanked Stat5a-Stat5b; K. Zhao and D. Schones for discussions; R.H. Schwartz, R. Spolski and Y. Rochman for comments; K. Cui for help in generating ChIP-Seq data; T.A. Waldmann (National Cancer Institute) for anti–human IL-2Rα (anti-Tac) and IL-2Rβ (Mikβ1); K. Murphy (Washington University St. Louis) for the pRV-Il12rb2 retroviral construct; and J. Zhu (National Institute of Allergy and Infectious Diseases of the US National Institutes of Health) for the pRV-Tbx21 construct. Supported by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (US National Institutes of Health).
Author information
Authors and Affiliations
Contributions
W.L. and J.-X.L. planned studies, did experiments, analyzed data and wrote the manuscript; L.W. did experiments; P.L. analyzed data and wrote the manuscript; and W.J.L. supervised the project, planned studies, analyzed data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1123 kb)
Rights and permissions
About this article
Cite this article
Liao, W., Lin, JX., Wang, L. et al. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol 12, 551–559 (2011). https://doi.org/10.1038/ni.2030
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2030
This article is cited by
-
Apoptotic tumor cell-derived microparticles loading Napabucasin inhibit CSCs and synergistic immune therapy
Journal of Nanobiotechnology (2023)
-
IL-6 prevents Th2 cell polarization by promoting SOCS3-dependent suppression of IL-2 signaling
Cellular & Molecular Immunology (2023)
-
Aging-associated HELIOS deficiency in naive CD4+ T cells alters chromatin remodeling and promotes effector cell responses
Nature Immunology (2023)
-
Strategies to therapeutically modulate cytokine action
Nature Reviews Drug Discovery (2023)
-
Ivermectin Protects Against Experimental Autoimmune Encephalomyelitis in Mice by Modulating the Th17/Treg Balance Involved in the IL-2/STAT5 Pathway
Inflammation (2023)