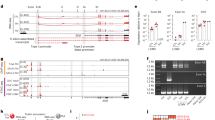
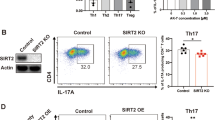
Abstract
Interleukin 2 (IL-2), a cytokine linked to human autoimmune disease, limits IL-17 production. Here we found that deletion of the gene encoding the transcription factor STAT3 in T cells abrogated IL-17 production and attenuated autoimmunity associated with IL-2 deficiency. Whereas STAT3 induced IL-17 and the transcription factor RORγt and inhibited the transcription factor Foxp3, IL-2 inhibited IL-17 independently of Foxp3 and RORγt. STAT3 and STAT5 bound to multiple common sites across the locus encoding IL-17. The induction of STAT5 binding by IL-2 was associated with less binding of STAT3 at these sites and the inhibition of associated active epigenetic marks. 'Titration' of the relative activation of STAT3 and STAT5 modulated the specification of cells to the IL-17-producing helper T cell (TH17 cell) subset. Thus, the balance rather than the absolute magnitude of these signals determined the propensity of cells to make a key inflammatory cytokine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Weaver, C.T. & Hatton, R.D. Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nat. Rev. Immunol. 9, 883–889 (2009).
Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Hsu, H.C. et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat. Immunol. 9, 166–175 (2008).
Veldhoen, M., Hocking, R.J., Atkins, C.J., Locksley, R.M. & Stockinger, B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Bettelli, E., Korn, T. & Kuchroo, V.K. Th17: the third member of the effector T cell trilogy. Curr. Opin. Immunol. 19, 652–657 (2007).
Harrington, L.E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005).
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010).
Chen, Z. et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl. Acad. Sci. USA 103, 8137–8142 (2006).
Durant, L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010).
Yang, X.O. et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282, 9358–9363 (2007).
Mathur, A.N. et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178, 4901–4907 (2007).
Laurence, A. et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007).
Harris, T.J. et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 179, 4313–4317 (2007).
Milner, J.D. et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776 (2008).
Cenit, M.C. et al. STAT3 locus in inflammatory bowel disease and multiple sclerosis susceptibility. Genes Immun. 11, 264–268 (2010).
Stumhofer, J.S. et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol. 7, 937–945 (2006).
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008).
Elias, K.M. et al. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood 111, 1013–1020 (2008).
Mucida, D. et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007).
Moriggl, R. et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity 10, 249–259 (1999).
Malek, T.R. The biology of interleukin-2. Annu. Rev. Immunol. 26, 453–479 (2008).
Sakaguchi, S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat. Immunol. 6, 345–352 (2005).
Yao, Z. et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375 (2007).
Diveu, C. et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. J. Immunol. 182, 5748–5756 (2009).
Wong, P.K. et al. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J. Clin. Invest. 116, 1571–1581 (2006).
Mukasa, R. et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32, 616–627 (2010).
Akimzhanov, A.M., Yang, X.O. & Dong, C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 282, 5969–5972 (2007).
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009).
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Nakajima, H., Brindle, P.K., Handa, M. & Ihle, J.N. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 20, 6836–6844 (2001).
Sadlack, B. et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75, 253–261 (1993).
Sadlack, B. et al. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur. J. Immunol. 25, 3053–3059 (1995).
Sharfe, N., Dadi, H.K., Shahar, M. & Roifman, C.M. Human immune disorder arising from mutation of the α chain of the interleukin-2 receptor. Proc. Natl. Acad. Sci. USA 94, 3168–3171 (1997).
Cohen, A.C. et al. Cutting edge: Decreased accumulation and regulatory function of CD4+CD25high T cells in human STAT5b deficiency. J. Immunol. 177, 2770–2774 (2006).
Hafler, D.A. et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357, 851–862 (2007).
Lowe, C.E. et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat. Genet. 39, 1074–1082 (2007).
Hoyer, K.K., Kuswanto, W.F., Gallo, E. & Abbas, A.K. Distinct roles of helper T-cell subsets in a systemic autoimmune disease. Blood 113, 389–395 (2009).
Park, J.H. et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 11, 257–264 (2010).
Wei, L. et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 (2010).
Esashi, E. et al. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity 28, 509–520 (2008).
Yu, M. et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682–695 (2009).
Walker, S.R. et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol. Cancer Res. 7, 966–976 (2009).
Rayman, J.B. et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16, 933–947 (2002).
Wells, J., Boyd, K.E., Fry, C.J., Bartley, S.M. & Farnham, P.J. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20, 5797–5807 (2000).
Takahashi, Y., Rayman, J.B. & Dynlacht, B.D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14, 804–816 (2000).
Saccani, S., Pantano, S. & Natoli, G. Modulation of NF-κB activity by exchange of dimers. Mol. Cell 11, 1563–1574 (2003).
Lee, C.K. et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity 17, 63–72 (2002).
Yang, X.O. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008).
Acknowledgements
We thank J. Simone, J. Lay and the National Institute of Arthritis, Musculoskeletal and Skin Diseases Laboratory Animal Care and Use Section staff for technical support; D. Levy (New York University) for mice with loxP-flanked Stat3 alleles; C. Dong (MD Anderson) for IL-17F–RFP reporter mice; W. Ouyang (Genentech) for monoclonal anti-IL-22; and S. Kuchen, F.C. Eberle and K. Tarbell for comments on the manuscript. Supported by the Intramural Research programs of National Institute of Arthritis, Musculoskeletal and Skin Diseases and National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
X.-P.Y. designed and did experiments, analyzed and wrote the manuscript; K.G. designed and did experiments and helped write the manuscript; S.M.S.-T. helped analyze gut lymphocytes; J.R.-C. examined the histopathology; J.Z., K.H. and J.R.G. provided mice and helped with experiments; Y.K. did ChIP-seq experiments; L.W., H.-W.S., Y.K. and G.V. analyzed data from the ChIP-seq and microarray experiments; J.J.O.'S. designed experiments, analyzed all acquired data and helped write the manuscript; and A.L. designed, did or interpreted all experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 1187 kb)
Rights and permissions
About this article
Cite this article
Yang, XP., Ghoreschi, K., Steward-Tharp, S. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 12, 247–254 (2011). https://doi.org/10.1038/ni.1995
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1995
This article is cited by
-
Immune modulation in malignant pleural effusion: from microenvironment to therapeutic implications
Cancer Cell International (2024)
-
IL-6 prevents Th2 cell polarization by promoting SOCS3-dependent suppression of IL-2 signaling
Cellular & Molecular Immunology (2023)
-
Peripheral Treg Levels and Transforming Growth Factor-β (TGFβ) in Patients with Psoriatic Arthritis: A Systematic Review Meta-Analysis
Advances in Therapy (2023)
-
The star target in SLE: IL-17
Inflammation Research (2023)
-
The role of Th17 cells in the pathogenesis and treatment of breast cancer
Cancer Cell International (2022)