Abstract
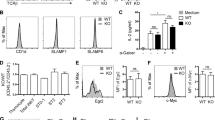
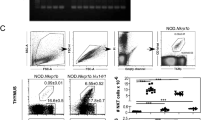
Type I invariant NKT cells (iNKT cells) are a subset of αβ T cells characterized by the expression of an invariant α-chain variable region 14–α-chain joining region 18 (Vα14Jα18) T cell antigen receptor (TCR) α-chain. The iNKT cells derive from CD4+CD8+ double-positive (DP) thymocytes, and their generation requires a long half-life of DP thymocytes to allow Vα14-Jα18 rearrangements, expression of glycolipid-loaded CD1d on DP thymocytes, and signaling through the signaling-activation molecule SLAM–adaptor SAP pathway. Here we show that the transcription factor c-Myb has a central role in priming DP thymocytes to enter the iNKT lineage by simultaneously regulating CD1d expression, the half-life of DP cells and expression of SLAMF1, SLAMF6 and SAP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Gapin, L., Matsuda, J.L., Surh, C.D. & Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2, 971–978 (2001).
Godfrey, D.I. & Berzins, S.P. Control points in NKT-cell development. Nat. Rev. Immunol. 7, 505–518 (2007).
Matsuda, J.L., Mallevaey, T., Scott-Browne, J. & Gapin, L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr. Opin. Immunol. 20, 358–368 (2008).
Bendelac, A., Savage, P.B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Kronenberg, M. & Engel, I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr. Opin. Immunol. 19, 186–193 (2007).
Krangel, M.S., Carabana, J., Abbarategui, I., Schlimgen, R. & Hawwari, A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunol. Rev. 200, 224–232 (2004).
Guo, J. et al. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat. Immunol. 3, 469–476 (2002).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Bezbradica, J.S., Hill, T., Stanic, A.K., Van Kaer, L. & Joyce, S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc. Natl. Acad. Sci. USA 102, 5114–5119 (2005).
Griewank, K. et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27, 751–762 (2007).
Borowski, C. & Bendelac, A. Signaling for NKT cell development: the SAP-FynT connection. J. Exp. Med. 201, 833–836 (2005).
Nichols, K.E. et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11, 340–345 (2005).
Pasquier, B. et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J. Exp. Med. 201, 695–701 (2005).
Chung, B., Aoukaty, A., Dutz, J., Terhorst, C. & Tan, R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J. Immunol. 174, 3153–3157 (2005).
Eberl, G., Lowin-Kropf, B. & MacDonald, H.R. Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J. Immunol. 163, 4091–4094 (1999).
Gapin, L. The making of NKT cells. Nat. Immunol. 9, 1009–1011 (2008).
Williams, J.A. et al. Regulation of thymic NKT cell development by the B7–CD28 costimulatory pathway. J. Immunol. 181, 907–917 (2008).
Felices, M. & Berg, L.J. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J. Immunol. 180, 3007–3018 (2008).
Matsuda, J.L. et al. Homeostasis of Vα14i NKT cells. Nat. Immunol. 3, 966–974 (2002).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Savage, A.K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 1–13 (2008).
Townsend, M.J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 20, 477–494 (2004).
Kim, P.J. et al. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 177, 6650–6659 (2006).
Dose, M. et al. Intrathymic proliferation wave essential for Vα14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA 106, 8641–8646 (2009).
Mucenski, M.L. et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–689 (1991).
Sakamoto, H. et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood 108, 896–903 (2006).
Greig, K.T., Carotta, S. & Nutt, S.L. Critical roles for c-Myb in hematopoietic progenitor cells. Semin. Immunol. 20, 247–256 (2008).
Bender, T.P., Kremer, C.S., Kraus, M., Buch, T. & Rajewsky, K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 5, 721–729 (2004).
Maurice, D., Hooper, J., Lang, G. & Weston, K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 26, 3629–3640 (2007).
Allen, R.D. III, Bender, T.P. & Siu, G. c-Myb is essential for early T cell development. Genes Dev. 13, 1073–1078 (1999).
Pearson, R. & Weston, K. c-Myb regulates the proliferation of immature thymocytes following β-selection. EMBO J. 19, 6112–6120 (2000).
Hernandez-Munain, C., Lauzurica, P. & Krangel, M.S. Regulation of T cell receptor δ gene rearrangement by c-Myb. J. Exp. Med. 183, 289–293 (1996).
Hsiang, Y.H., Goldman, J.P. & Raulet, D.H. The role of c-Myb or a related factor in regulating the T cell receptor γ gene enhancer. J. Immunol. 154, 5195–5204 (1995).
Liu, Y. et al. A modified α-galactosyl ceramide for staining and stimulating natural killer T cells. J. Immunol. Methods 312, 34–39 (2006).
Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Chao, D.T. & Korsmeyer, S.J. BCL-XL-regulated apoptosis in T cell development. Int. Immunol. 9, 1375–1384 (1997).
Cui, J. et al. Requirement for Vα14 NKT cells in IL-12-mediated rejection of tumors. Science 278, 1623–1626 (1997).
Hossain, M.Z., Yu, Q., Xu, M. & Sen, J.M. ICAT expression disrupts β-catenin-TCF interactions and impairs survival of thymocytes and activated mature T cells. Int. Immunol. 20, 925–935 (2008).
Xie, H., Huang, Z., Sadim, M.S. & Sun, Z. Stabilized β-catenin extends thymocyte survival by up-regulating Bcl-xL. J. Immunol. 175, 7981–7988 (2005).
Wandstrat, A.E. et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 21, 769–780 (2004).
Jordan, M.A., Fletcher, J.M., Pellicci, D. & Baxter, A.G. Slamf1, the NKT cell control gene Nkt1. J. Immunol. 178, 1618–1627 (2007).
Esteban, L.M. et al. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J. Immunol. 171, 2873–2878 (2003).
Berzins, S.P., Cochrane, A.D., Pellicci, D.G., Smyth, M.J. & Godfrey, D. I. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur. J. Immunol. 35, 1399–1407 (2005).
Morel, L., Yu, Y., Blenman, K.R., Caldwell, R.A. & Wakeland, E.K. Production of congenic mouse strains carrying genomic intervals containing SLE-susceptibility genes derived from the SLE-prone NZM2410 strain. Mamm. Genome 7, 335–339 (1996).
Chen, Y.H., Chiu, N.M., Mandal, M., Wang, N. & Wang, C.R. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 6, 459–467 (1997).
Hernandez-Hoyos, G. & Alberola-Ila, J. Analysis of T-cell development by using short interfering RNA to knock down protein expression. Methods Enzymol. 392, 199–217 (2005).
Lauritsen, J.P. et al. Egr2 is required for Bcl-2 induction during positive selection. J. Immunol. 181, 7778–7785 (2008).
Acknowledgements
We thank A. Bendelac (Howard Hughes Medical Institute) for the Vα14-transgenic mice; A. Veillette (Clinical Research Institute, Montreal) for the antibody to SAP; X.-H. Sun (Oklahoma Medical Research Foundation) for the Mig-RORγt vector; M. Lang (University of Oklahoma Health Science Center) for CD1d-deficient- and Jα18-deficient- bone marrow; the National Institutes of Health Tetramer Facility for the CD1d-PBS57 tetramer; and S. Kovats for discussions. Supported by the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (AI059302 to J.A.-I.), the American Heart Association (0855002F to J.A.-I.), the National Cancer Institute of the National Institutes of Health (CA85842 to T.B.P.) and the Robert R. Wagner Fellowship Fund (J.Y.).
Author information
Authors and Affiliations
Contributions
T.H. designed the study, did experiments, analyzed data and wrote the manuscript; A.S. did experiments; J.Y and T.P.B. provided experimental mice, data and discussions; and J.A.-I. designed the study, did experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Note (PDF 1101 kb)
Rights and permissions
About this article
Cite this article
Hu, T., Simmons, A., Yuan, J. et al. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol 11, 435–441 (2010). https://doi.org/10.1038/ni.1865
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1865
This article is cited by
-
Chromatin organizer SATB1 controls the cell identity of CD4+ CD8+ double-positive thymocytes by regulating the activity of super-enhancers
Nature Communications (2022)
-
SRSF1 plays a critical role in invariant natural killer T cell development and function
Cellular & Molecular Immunology (2021)
-
microRNA dynamic expression regulates invariant NKT cells
Cellular and Molecular Life Sciences (2021)
-
Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge
Nature Reviews Immunology (2020)
-
Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells
Nature Communications (2020)