Abstract
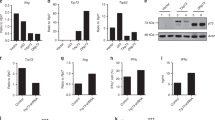
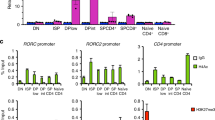
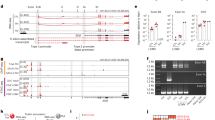
T helper type 2 (TH2) bias, which is the propensity of naive CD4+ T cells to differentiate into interleukin 4 (IL-4)-secreting TH2 cells, is a genetic trait that affects susceptibility to infectious, autoimmune and allergic diseases. TH2 bias correlates with the amount of IL-4 initially secreted by newly activated helper T cells that feeds back positively through the pathway of the IL-4 receptor and the transcription factors STAT6 and GATA-3 to drive TH2 development. Here we identify Mina, a member of the jumonji C (JmjC) protein family, as a genetic determinant of TH2 bias. Mina specifically bound to and repressed the Il4 promoter. Mina overexpression in transgenic mice impaired Il4 expression, whereas its knockdown in primary CD4+ T cells led to Il4 derepression. Our findings collectively provide mechanistic insight into an Il4-regulatory pathway that controls helper T cell differentiation and genetic variation in TH2 bias.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
05 July 2009
In the version of this article initially published online, the second corresponding author initials were incorrect. The correct initials are “M.Ku.” The error has been corrected for the print, PDF and HTML versions of this article.
References
Mosmann, T.R. & Coffman, R.L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7, 145–173 (1989).
Rook, G.A. Th2 cytokines in susceptibility to tuberculosis. Curr. Mol. Med. 7, 327–337 (2007).
Wilson, K.T. & Fantry, G.T. Pathogenesis of Helicobacter pylori infection. Curr. Opin. Gastroenterol. 15, 66–71 (1999).
Umetsu, D.T. & Dekruyff, R.H. Immune dysregulation in asthma. Curr. Opin. Immunol. 18, 727–732 (2006).
Crane, I.J. & Forrester, J.V. Th1 and Th2 lymphocytes in autoimmune disease. Crit. Rev. Immunol. 25, 75–102 (2005).
Ngoc, P.L., Gold, D.R., Tzianabos, A.O., Weiss, S.T. & Celedon, J.C. Cytokines, allergy, and asthma. Curr. Opin. Allergy Clin. Immunol. 5, 161–166 (2005).
Pearce, E.J. et al. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol. Rev. 201, 117–126 (2004).
Maizels, R.M. et al. Helminth parasites–masters of regulation. Immunol. Rev. 201, 89–116 (2004).
Wynn, T.A. Fibrotic disease and the TH1/TH2 paradigm. Nat. Rev. Immunol. 4, 583–594 (2004).
Swain, S.L., Weinberg, A.D., English, M. & Huston, G. IL-4 directs the development of Th2-like helper effectors. J. Immunol. 145, 3796–3806 (1990).
Mowen, K.A. & Glimcher, L.H. Signaling pathways in Th2 development. Immunol. Rev. 202, 203–222 (2004).
Zhu, J., Yamane, H., Cote-Sierra, J., Guo, L. & Paul, W.E. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 16, 3–10 (2006).
Le Gros, G., Ben-Sasson, S.Z., Seder, R., Finkelman, F.D. & Paul, W.E. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4- producing cells. J. Exp. Med. 172, 921–929 (1990).
Bix, M., Wang, Z.E., Thiel, B., Schork, N.J. & Locksley, R.M. Genetic regulation of commitment to interleukin 4 production by a CD4+ T cell-intrinsic mechanism. J. Exp. Med. 188, 2289–2299 (1998).
Noben-Trauth, N., Hu-Li, J. & Paul, W.E. IL-4 secreted from individual naive CD4+ T cells acts in an autocrine manner to induce Th2 differentiation. Eur. J. Immunol. 32, 1428–1433 (2002).
Yagi, R. et al. The IL-4 production capability of different strains of naive CD4+ T cells controls the direction of the TH cell response. Int. Immunol. 14, 1–11 (2002).
Murphy, K.M. & Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002).
Liu, Z. et al. Requirements for the development of IL-4-producing T cells during intestinal nematode infections: what it takes to make a Th2 cell in vivo. Immunol. Rev. 201, 57–74 (2004).
Hsieh, C.S., Macatonia, S.E., O'Garra, A. & Murphy, K.M. T cell genetic background determines default T helper phenotype development in vitro. J. Exp. Med. 181, 713–721 (1995).
Yagi, J. et al. Genetic background influences Th cell differentiation by controlling the capacity for IL-2-induced IL-4 production by naive CD4+ T cells. Int. Immunol. 18, 1681–1690 (2006).
Reiner, S.L. & Locksley, R.M. The regulation of immunity to Leishmania major. Annu. Rev. Immunol. 13, 151–177 (1995).
Yamashita, M., Onodera, A. & Nakayama, T. Immune mechanisms of allergic airway disease: regulation by transcription factors. Crit. Rev. Immunol. 27, 539–546 (2007).
Kuroda, E., Sugiura, T., Zeki, K., Yoshida, Y. & Yamashita, U. Sensitivity difference to the suppressive effect of prostaglandin E2 among mouse strains: a possible mechanism to polarize Th2 type response in BALB/c mice. J. Immunol. 164, 2386–2395 (2000).
Gorham, J.D. et al. Genetic mapping of a murine locus controlling development of T helper 1/T helper 2 type responses. Proc. Natl. Acad. Sci. USA 93, 12467–12472 (1996).
Guler, M.L. et al. Tpm1, a locus controlling IL-12 responsiveness, acts by a cell-autonomous mechanism. J. Immunol. 162, 1339–1347 (1999).
Kosarova, M., Havelkova, H., Krulova, M., Demant, P. & Lipoldova, M. The production of two Th2 cytokines, interleukin-4 and interleukin-10, is controlled independently by locus Cypr1 and by loci Cypr2 and Cypr3, respectively. Immunogenetics 49, 134–141 (1999).
Zhang, F. et al. A murine locus on chromosome 18 controls NKT cell homeostasis and Th cell differentiation. J. Immunol. 171, 4613–4620 (2003).
Baguet, A., Epler, J., Wen, K.W. & Bix, M. A Leishmania major response locus identified by interval-specific congenic mapping of a T helper type 2 cell bias-controlling quantitative trait locus. J. Exp. Med. 200, 1605–1612 (2004).
Choi, P. et al. Linkage analysis of the genetic determinants of T-cell IL-4 secretion, and identification of Flj20274 as a putative candidate gene. Genes Immun. 6, 290–297 (2005).
Eppig, J.T., Blake, J.A., Bult, C.J., Kadin, J.A. & Richardson, J.E. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 35, D630–D637 (2007).
Lipson, K.E. & Baserga, R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl. Acad. Sci. USA 86, 9774–9777 (1989).
Tokoyoda, K., Takemoto, Y., Nakayama, T., Arai, T. & Kubo, M. Synergism between the calmodulin-binding and autoinhibitory domains on calcineurin is essential for the induction of their phosphatase activity. J. Biol. Chem. 275, 11728–11734 (2000).
Bruhn, K.W., Nelms, K., Boulay, J.L., Paul, W.E. & Lenardo, M.J. Molecular dissection of the mouse interleukin-4 promoter. Proc. Natl. Acad. Sci. USA (1993).
Chuvpilo, S. et al. Multiple closely-linked NFAT/octamer and HMG I(Y) binding sites are part of the interleukin-4 promoter. Nucleic Acids Res. 21, 5694–5704 (1993).
Farrar, J.J. et al. Thymoma production of T cell growth factor (Interleukin 2). J. Immunol. 125, 2555–2558 (1980).
Iritani, B.M., Forbush, K.A., Farrar, M.A. & Perlmutter, R.M. Control of B cell development by Ras-mediated activation of Raf. EMBO J. 16, 7019–7031 (1997).
Klose, R.J., Kallin, E.M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006).
Chang, B., Chen, Y., Zhao, Y. & Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007).
Dann, C.E., Bruick, R.K. & Deisenhofer, J. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl. Acad. Sci. USA 99, 15351–15356 (2002).
Elkins, J.M. et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278, 1802–1806 (2003).
Baguet, A. & Bix, M. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc. Natl. Acad. Sci. USA 101, 11410–11415 (2004).
Tanaka, S. et al. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity 24, 689–701 (2006).
Amsen, D. et al. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117, 515–526 (2004).
Mowen, K.A., Schurter, B.T., Fathman, J.W., David, M. & Glimcher, L.H. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15, 559–571 (2004).
Nishimune, Y., Ohta, K., Tanaka, H., Iida, H. & Inokuchi, T. Expression of Mina53, a product of a Myc target gene in mouse testis. Int. J. Androl. 29, 323–330 (2006).
Tsuneoka, M. et al. Mina53 as a potential prognostic factor for esophageal squamous cell carcinoma. Clin. Cancer Res. 10, 7347–7356 (2004).
Teye, K. et al. Increased expression of a Myc target gene Mina53 in human colon cancer. Am. J. Pathol. 164, 205–216 (2004).
Tsuneoka, M., Koda, Y., Soejima, M., Teye, K. & Kimura, H. A novel myc target gene, mina53, that is involved in cell proliferation. J. Biol. Chem. 277, 35450–35459 (2002).
Dubchak, I. et al. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304–1306 (2000).
Hijikata, A. et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics 23, 2934–2941 (2007).
Seki, N. et al. IL-4-induced GATA-3 expression is a time-restricted instruction switch for Th2 cell differentiation. J. Immunol. 172, 6158–6166 (2004).
Rooney, J.W., Sun, Y.L., Glimcher, L.H. & Hoey, T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol. 15, 6299–6310 (1995).
Ragione, F.D. et al. p21Cip1 gene expression is modulated by Egr1: a novel regulatory mechanism involved in the resveratrol antiproliferative effect. J. Biol. Chem. 278, 23360–23368 (2003).
Koyanagi, M. et al. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in TH1 cells. J. Biol. Chem. 280, 31470–31477 (2005).
Weinmann, A.S., Bartley, S.M., Zhang, T., Zhang, M.Q. & Farnham, P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21, 6820–6832 (2001).
Attar, R.M., Macdonald-Bravo, H., Raventos-Suarez, C., Durham, S.K. & Bravo, R. Expression of constitutively active IκBβ in T cells of transgenic mice: persistent NF-κB activity is required for T-cell immune responses. Mol. Cell. Biol. 18, 477–487 (1998).
Summerton, J.E. Endo-Porter: a novel reagent for safe, effective delivery of substances into cells. Ann. NY Acad. Sci. 1058, 62–75 (2005).
Acknowledgements
We thank R. Abe (Science University of Tokyo) for hybridoma PV-1; J. Allison (Memorial Sloan Kettering Cancer) for hybridoma 37N51.1; G. Trinchieri (US National Insitutes of Health) for hybridoma C17.8; A. Matsuno, M. Nakamura, M. Natsume, J. Epler, Y. Zhang, N. Li, S. Brown and R. Cross for technical help; J. Partridge for discussions; and D. Green, H. Beere and J. Kang (U. Mass. Medical School) for comments on the manuscript. Supported by the Cancer Research Institute (M.B.), the Burroughs Wellcome Fund (M.B.), American Lebanese Syrian Associated Charities (M.B.), the RIKEN Research Center for Allergy and Immunology International Collaboration Award Program (M.B. and M.Ku.), a Grant-in-Aid for Scientific Research (B) (M.Ku.), a Grant-in-Aid for Scientific Research on Priority Areas of the Ministry of Education, Culture, Sports, Science, and Technology (Japan) (M.Ku.), the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (M.Ku.) and the US National Institutes of Health (AI048636 to M.B.).
Author information
Authors and Affiliations
Contributions
M.O. and M.V.S. did the experiments; L.C. did the Mina transcriptional analysis and Figures 2b and 3b and Supplementary Figure 2; M.Ka. did the Mina immunoblots; X.S. analyzed the C16D2/8D mice; Y.S. did the transgenic experiments; Y.S. and L.C. maintained the mouse colonies; O.O., H.K. and A.H. did the expression profiling; M.O., M.V.S., M.Ku and M.B. designed and conceptualized the research and analyzed the data; and M.B. prepared the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 958 kb)
Rights and permissions
About this article
Cite this article
Okamoto, M., Van Stry, M., Chung, L. et al. Mina, an Il4 repressor, controls T helper type 2 bias. Nat Immunol 10, 872–879 (2009). https://doi.org/10.1038/ni.1747
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1747
This article is cited by
-
Phylogenetic and genomic analyses of the ribosomal oxygenases Riox1 (No66) and Riox2 (Mina53) provide new insights into their evolution
BMC Evolutionary Biology (2018)
-
The emerging roles of ribosomal histidyl hydroxylases in cell biology, physiology and disease
Cellular and Molecular Life Sciences (2018)
-
Dysfunction of IKZF1/MYC/MDIG axis contributes to liver cancer progression through regulating H3K9me3/p21 activity
Cell Death & Disease (2017)
-
Development and validation of an Onchocerca ochengi microfilarial hamster model for onchocerciasis drug screens
BMC Infectious Diseases (2016)
-
T-bet is a key modulator of IL-23-driven pathogenic CD4+ T cell responses in the intestine
Nature Communications (2016)