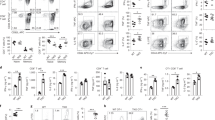
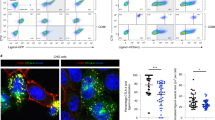
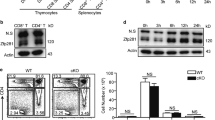
Abstract
Nedd4 and Itch are E3 ubiquitin ligases that ubiquitinate similar targets in vitro and thus are thought to function similarly. T cells lacking Itch show spontaneous activation and T helper type 2 polarization. To test whether loss of Nedd4 affects T cells in the same way, we generated Nedd4+/+ and Nedd4−/− fetal liver chimeras. Nedd4−/− T cells developed normally but proliferated less, produced less interleukin 2 and provided inadequate help to B cells. Nedd4−/− T cells contained more of the E3 ubiquitin ligase Cbl-b, and Nedd4 was required for polyubiquitination of Cbl-b induced by CD28 costimulation. Our data demonstrate that Nedd4 promotes the conversion of naive T cells into activated T cells. We propose that Nedd4 and Itch ubiquitinate distinct target proteins in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Ciechanover, A., Heller, H., Elias, S., Haas, A.L. & Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 77, 1365–1368 (1980).
Ciechanover, A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 12, 1178–1190 (2005).
Liu, Y.C. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 22, 81–127 (2004).
Fang, D. et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3, 281–287 (2002).
Oliver, P.M. et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940 (2006).
Heissmeyer, V. et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5, 255–265 (2004).
Babu, S., Blauvelt, C.P., Kumaraswami, V. & Nutman, T.B. Regulatory networks induced by live parasites impair both Th1 and Th2 pathways in patent lymphatic filariasis: implications for parasite persistence. J. Immunol. 176, 3248–3256 (2006).
Perry, W.L. et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18, 143–146 (1998).
Liu, Y.C. The E3 ubiquitin ligase Itch in T cell activation, differentiation, and tolerance. Semin. Immunol. 19, 197–205 (2007).
Magnifico, A. et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 278, 43169–43177 (2003).
Scharschmidt, E., Wegener, E., Heissmeyer, V., Rao, A. & Krappmann, D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-κB signaling. Mol. Cell. Biol. 24, 3860–3873 (2004).
Bonnevier, J.L., Zhang, R. & Mueller, D.L. E3 ubiquitin ligases and their control of T cell autoreactivity. Arthritis Res. Ther. 7, 233–242 (2005).
Mueller, D.L. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 5, 883–890 (2004).
Ingham, R.J. et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 25, 7092–7106 (2005).
Stryke, D. et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 31, 278–281 (2003).
Rees, W. et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4+ T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA 96, 9781–9786 (1999).
Grewal, I.S. & Flavell, R.A. The role of CD40 ligand in costimulation and T-cell activation. Immunol. Rev. 153, 85–106 (1996).
Powell, J.D., Ragheb, J.A., Kitagawa-Sakakida, S. & Schwartz, R.H. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol. Rev. 165, 287–300 (1998).
Umlauf, S.W., Beverly, B., Lantz, O. & Schwartz, R.H. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol. Cell. Biol. 15, 3197–3205 (1995).
Green, J.M. et al. Absence of B7-dependent responses in CD28-deficient mice. Immunity 1, 501–508 (1994).
Lucas, P.J., Negishi, I., Nakayama, K., Fields, L.E. & Loh, D.Y. Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response. J. Immunol. 154, 5757–5768 (1995).
Shahinian, A. et al. Differential T cell costimulatory requirements in CD28-deficient mice. Science 261, 609–612 (1993).
Trotman, L.C. et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156 (2007).
Wang, X. et al. NEDD4 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007).
Leykauf, K. et al. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J. Cell Sci. 119, 3634–3642 (2006).
Seminario, M.C. & Wange, R.L. Lipid phosphatases in the regulation of T cell activation: living up to their PTEN-tial. Immunol. Rev. 192, 80–97 (2003).
Xu, Z., Stokoe, D., Kane, L.P. & Weiss, A. The inducible expression of the tumor suppressor gene PTEN promotes apoptosis and decreases cell size by inhibiting the PI3K/Akt pathway in Jurkat T cells. Cell Growth Differ. 13, 285–296 (2002).
Buckler, J.L., Walsh, P.T., Porrett, P.M., Choi, Y. & Turka, L.A. Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J. Immunol. 177, 4262–4266 (2006).
Zhang, J. et al. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J. Immunol. 169, 2236–2240 (2002).
Kim, H.T. et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 (2007).
Levy, F. et al. Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol. Biol. Cell 16, 1777–1787 (2005).
Murdaca, J. et al. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J. Biol. Chem. 279, 26754–26761 (2004).
Vecchione, A., Marchese, A., Henry, P., Rotin, D. & Morrione, A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol. Cell. Biol. 23, 3363–3372 (2003).
Sakata, T. et al. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr. Biol. 14, 2228–2236 (2004).
Jennings, M.D., Blankley, R.T., Baron, M., Golovanov, A.P. & Avis, J.M. Specificity and autoregulation of Notch binding by tandem WW domains in suppressor of Deltex. J. Biol. Chem. 282, 29032–29042 (2007).
Chiang, Y.J. et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403, 216–220 (2000).
Di Cristofano, A. et al. Impaired Fas response and autoimmunity in Pten+/− mice. Science 285, 2122–2125 (1999).
Donovan, J.A., Wange, R.L., Langdon, W.Y. & Samelson, L.E. The protein product of the c–cbl protooncogene is the 120-kDa tyrosine-phosphorylated protein in Jurkat cells activated via the T cell antigen receptor. J. Biol. Chem. 269, 22921–22924 (1994).
Gaide, O. et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat. Immunol. 3, 836–843 (2002).
Robey, E. et al. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 87, 483–492 (1996).
Thome, M. & Tschopp, J. TCR-induced NF-κB activation: a crucial role for Carma1, Bcl10 and MALT1. Trends Immunol. 24, 419–424 (2003).
Villalba, M. et al. Protein kinase cθ cooperates with calcineurin to induce Fas ligand expression during activation-induced T cell death. J. Immunol. 163, 5813–5819 (1999).
Washburn, T. et al. Notch activity influences the αβ versus γδ T cell lineage decision. Cell 88, 833–843 (1997).
McDonald, F.J. et al. Disruption of the β subunit of the epithelial Na+ channel in mice: hyperkalemia and neonatal death associated with a pseudohypoaldosteronism phenotype. Proc. Natl. Acad. Sci. USA 96, 1727–1731 (1999).
Cao, X.R. et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. (in the press).
McCormack, J.E., Kappler, J. & Marrack, P. Stimulation with specific antigen can block superantigen-mediated deletion of T cells in vivo. Proc. Natl. Acad. Sci. USA 91, 2086–2090 (1994).
Acknowledgements
We thank R. Schwartz (National Institute of Allergy and Infectious Diseases) for Cbl-b-deficient mice; M. Shaffer for help with siRNA experiments; and J. Loomis for sorting cells.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 207 kb)
Rights and permissions
About this article
Cite this article
Yang, B., Gay, D., MacLeod, M. et al. Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat Immunol 9, 1356–1363 (2008). https://doi.org/10.1038/ni.1670
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1670
This article is cited by
-
NEDD4 ameliorates myocardial reperfusion injury by preventing macrophages pyroptosis
Cell Communication and Signaling (2023)
-
Loss of NEDD4 causes complete XY gonadal sex reversal in mice
Cell Death & Disease (2022)
-
Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4+ T cells
Nature Immunology (2019)
-
The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR
Nature Immunology (2018)
-
Development of an efficient screening system to identify novel bone metabolism-related genes using the exchangeable gene trap mutagenesis mouse models
Scientific Reports (2017)