Abstract
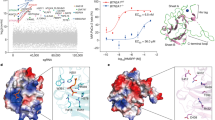
Protein phosphorylation generates a source of phosphopeptides that are presented by major histocompatibility complex class I molecules and recognized by T cells. As deregulated phosphorylation is a hallmark of malignant transformation, the differential display of phosphopeptides on cancer cells provides an immunological signature of 'transformed self'. Here we demonstrate that phosphorylation can considerably increase peptide binding affinity for HLA-A2. To understand this, we solved crystal structures of four phosphopeptide–HLA-A2 complexes. These identified a novel peptide-binding motif centered on a solvent-exposed phosphate anchor. Our findings indicate that deregulated phosphorylation can create neoantigens by promoting binding to major histocompatibility complex molecules or by affecting the antigenic identity of presented epitopes. These results highlight the potential of phosphopeptides as novel targets for cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rock, K.L. & Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17, 739–779 (1999).
Engelhard, V.H., Altrich-Vanlith, M., Ostankovitch, M. & Zarling, A.L. Post-translational modifications of naturally processed MHC-binding epitopes. Curr. Opin. Immunol. 18, 92–97 (2006).
Zarling, A.L. et al. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 192, 1755–1762 (2000).
Zarling, A.L. et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. USA 103, 14889–14894 (2006).
Andersen, M.H. et al. Phosphorylated peptides can be transported by TAP molecules, presented by class I MHC molecules, and recognized by phosphopeptide-specific CTL. J. Immunol. 163, 3812–3818 (1999).
Harper, J.W. A phosphorylation-driven ubiquitination switch for cell-cycle control. Trends Cell Biol. 12, 104–107 (2002).
Koepp, D.M., Harper, J.W. & Elledge, S.J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97, 431–434 (1999).
Reed, S.I. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4, 855–864 (2003).
Ang, X.L. & Wade Harper, J. SCF-mediated protein degradation and cell cycle control. Oncogene 24, 2860–2870 (2005).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Wilkinson, K.D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11, 141–148 (2000).
Princiotta, M.F. et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18, 343–354 (2003).
Blume-Jensen, P. & Hunter, T. Oncogenic kinase signalling. Nature 411, 355–365 (2001).
Evan, G.I. & Vousden, K.H. Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 (2001).
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Fecher, L.A., Cummings, S.D., Keefe, M.J. & Alani, R.M. Toward a molecular classification of melanoma. J. Clin. Oncol. 25, 1606–1620 (2007).
Easty, D.J. & Bennett, D.C. Protein tyrosine kinases in malignant melanoma. Melanoma Res. 10, 401–411 (2000).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Diella, F. et al. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics 5, 79 (2004).
Hornbeck, P.V., Chabra, I., Kornhauser, J.M., Skrzypek, E. & Zhang, B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 (2004).
Bjorkman, P.J. et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512 (1987).
Madden, D.R., Garboczi, D.N. & Wiley, D.C. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708 (1993).
Madden, D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 (1995).
Ruppert, J. et al. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 74, 929–937 (1993).
Bouvier, M. & Wiley, D.C. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science 265, 398–402 (1994).
Fersht, A.R. et al. Hydrogen bonding and biological specificity analysed by protein engineering. Nature 314, 235–238 (1985).
Sette, A. et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153, 5586–5592 (1994).
Hendrickson, R.C., Skipper, J.C., Shabanowitz, J., Slingluff, C.L., Jr. & Engelhard, V.H . in Immunology Methods Manual: The Comprehensive Sourcebook of Techniques (ed. Lefkovits, I.) 605–623 (Academic, New York, 1996).
Martin, S.E., Shabanowitz, J., Hunt, D.F. & Marto, J.A. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 72, 4266–4274 (2000).
Sidney, J. et al. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum. Immunol. 62, 1200–1216 (2001).
Gulukota, K., Sidney, J., Sette, A. & DeLisi, C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 267, 1258–1267 (1997).
Garboczi, D.N., Hung, D.T. & Wiley, D.C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA 89, 3429–3433 (1992).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Collaborative Computational Project No 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Brünger, A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Fenn, T.D., Ringe, D. & Petsko, G.A. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Cryst. 36, 944–947 (2003).
Guex, N. & Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct. Funct. Genet. 11, 281–296 (1991).
Acknowledgements
We thank S. White and K. Fütterer for help with X-ray data collection, and P. Hornbeck (Cell Signaling Technology; PhosphoSites) and F. Diella (European Molecular Biology Laboratory, Heidelberg; Phospho.ELM) for data sets of identified human phosphorylation sites. Supported by US Public Health Service (AI20963 to V.H.E. and AI33993 to D.F.H.), the Medical Research Council (B.E.W. and M.C.), the Wellcome Trust (B.E.W.), the Biotechnology and Biological Sciences Research Council (F.M.) and the Sidney Kimmel Foundation (A.L.Z.).
Author information
Authors and Affiliations
Contributions
F.M., M.C., A.L.Z., M.S., V.H.E. and B.E.W. did the experiments and/or analyzed the data; G.A.B.-W., J.S. and D.F.H. contributed to the mass spectrometry data and analysis; B.E.W, M.C. and V.H.E. designed the study; and B.E.W. and V.H.E. prepared the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Tables 1–8 (PDF 5306 kb)
Rights and permissions
About this article
Cite this article
Mohammed, F., Cobbold, M., Zarling, A. et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol 9, 1236–1243 (2008). https://doi.org/10.1038/ni.1660
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1660
This article is cited by
-
Molecular mechanism of phosphopeptide neoantigen immunogenicity
Nature Communications (2023)
-
Murine xenograft bioreactors for human immunopeptidome discovery
Scientific Reports (2019)
-
Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics
Nature Protocols (2019)
-
T cell receptor cross-reactivity expanded by dramatic peptide–MHC adaptability
Nature Chemical Biology (2018)
-
Analysis of the affinity of influenza A virus protein epitopes for swine MHC I by a modified in vitro refolding method indicated cross-reactivity between swine and human MHC I specificities
Immunogenetics (2018)