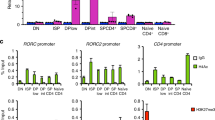
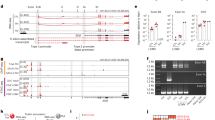
Abstract
T helper type 2 (TH2) cells are essential for humoral immunity and host defense. Interleukin 4 (IL-4) drives TH2 differentiation and IL-2 augments the accessibility of Il4 chromatin. Here we demonstrate that IL-2, by inducing binding of STAT5 to the Il4ra locus, which encodes IL-4 receptor α-chain (IL-4Rα), was essential for inducing and maintaining IL-4Rα expression. Although IL-4 induced IL-4Rα expression, T cell receptor–induced IL-4Rα expression was normal in Il4−/− cells but was much lower in Il2−/− cells. Notably, forced IL-4Rα expression restored the TH2 differentiation of Il2−/− cells. Moreover, genome-wide mapping by chromatin immunoprecipitation coupled with sequencing showed broad interaction of the transcription factors STAT5A and STAT5B with genes associated with TH2 differentiation. Our results identify a previously unappreciated function for IL-2 in 'priming' T cells for TH2 differentiation and in maintaining the expression of Il4ra and other genes in TH2-committed cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Accession codes
References
Murphy, K.M. & Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002).
Szabo, S.J., Sullivan, B.M., Peng, S.L. & Glimcher, L.H. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 21, 713–758 (2003).
Ansel, K.M., Djuretic, I., Tanasa, B. & Rao, A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24, 607–656 (2006).
Weaver, C.T., Hatton, R.D., Mangan, P.R. & Harrington, L.E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007).
Yang, X.O. et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 282, 9358–9363 (2007).
Spolski, R. & Leonard, W.J. Interleukin-21: basic biology and implications for cancer and autoimmunity. Annu. Rev. Immunol. 26, 57–80 (2007).
Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Rogge, L. et al. Transcript imaging of the development of human T helper cells using oligonucleotide arrays. Nat. Genet. 25, 96–101 (2000).
Zhu, J., Guo, L., Watson, C.J., Hu-Li, J. & Paul, W.E. Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J. Immunol. 166, 7276–7281 (2001).
Shimoda, K. et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 380, 630–633 (1996).
Kaplan, M.H., Schindler, U., Smiley, S.T. & Grusby, M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4, 313–319 (1996).
Zhu, J., Cote-Sierra, J., Guo, L. & Paul, W.E. Stat5 activation plays a critical role in Th2 differentiation. Immunity 19, 739–748 (2003).
Coffman, R.L. & von der Weid, T. Multiple pathways for the initiation of T helper 2 (Th2) responses. J. Exp. Med. 185, 373–375 (1997).
Voehringer, D., Reese, T.A., Huang, X., Shinkai, K. & Locksley, R.M. Type 2 immunity is controlled by IL-4/IL-13 expression in hematopoietic non-eosinophil cells of the innate immune system. J. Exp. Med. 203, 1435–1446 (2006).
Hilton, D.J. et al. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc. Natl. Acad. Sci. USA 93, 497–501 (1996).
Aman, M.J. et al. cDNA cloning and characterization of the human interleukin 13 receptor α chain. J. Biol. Chem. 271, 29265–29270 (1996).
Lin, J.X. et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 2, 331–339 (1995).
Nelms, K., Keegan, A.D., Zamorano, J., Ryan, J.J. & Paul, W.E. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17, 701–738 (1999).
Ohara, J. & Paul, W.E. Up-regulation of interleukin 4/B-cell stimulatory factor 1 receptor expression. Proc. Natl. Acad. Sci. USA 85, 8221–8225 (1988).
Kovanen, P.E. et al. Global analysis of IL-2 target genes: identification of chromosomal clusters of expressed genes. Int. Immunol. 17, 1009–1021 (2005).
Xue, H.H. et al. IL-2 negatively regulates IL-7 receptor alpha chain expression in activated T lymphocytes. Proc. Natl. Acad. Sci. USA 99, 13759–13764 (2002).
Renz, H., Domenico, J. & Gelfand, E.W. IL-4-dependent up-regulation of IL-4 receptor expression in murine T and B cells. J. Immunol. 146, 3049–3055 (1991).
Dokter, W.H. et al. Interleukin-4 (IL-4) receptor expression on human T cells is affected by different intracellular signaling pathways and by IL-4 at transcriptional and posttranscriptional level. Blood 80, 2721–2728 (1992).
Dautry, F., Weil, D., Yu, J. & Dautry-Varsat, A. Regulation of Pim and Myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J. Biol. Chem. 263, 17615–17620 (1988).
Zhu, J. et al. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 16, 733–744 (2002).
Cote-Sierra, J. et al. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 101, 3880–3885 (2004).
Leonard, W.J. & O'Shea, J.J. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16, 293–322 (1998).
Kelly, J.A. et al. Stat5 synergizes with T cell receptor/antigen stimulation in the development of lymphoblastic lymphoma. J. Exp. Med. 198, 79–89 (2003).
Cui, Y. et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24, 8037–8047 (2004).
Soldaini, E. et al. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell. Biol. 20, 389–401 (2000).
Auernhammer, C.J., Bousquet, C. & Melmed, S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl. Acad. Sci. USA 96, 6964–6969 (1999).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Johnson, D.S., Mortazavi, A., Myers, R.M. & Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007).
Kagami, S. et al. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood 97, 2358–2365 (2001).
Barner, M., Mohrs, M., Brombacher, F. & Kopf, M. Differences between IL-4Rα-deficient and IL-4-deficient mice reveal a role for IL-13 in the regulation of Th2 responses. Curr. Biol. 8, 669–672 (1998).
Noben-Trauth, N. et al. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc. Natl. Acad. Sci. USA 94, 10838–10843 (1997).
Pernis, A.B. & Rothman, P.B. JAK-STAT signaling in asthma. J. Clin. Invest. 109, 1279–1283 (2002).
Wynn, T.A. IL-13 effector functions. Annu. Rev. Immunol. 21, 425–456 (2003).
Kim, H.P., Imbert, J. & Leonard, W.J. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 17, 349–366 (2006).
Kelly, J. et al. A role for Stat5 in CD8+ T cell homeostasis. J. Immunol. 170, 210–217 (2003).
Kim, H.P., Kelly, J. & Leonard, W.J. The basis for IL-2-induced IL-2 receptor α chain gene regulation: importance of two widely separated IL-2 response elements. Immunity 15, 159–172 (2001).
Acknowledgements
We thank P. Ryan (National Heart, Lung and Blood Institute, National Institutes of Health) for Il4−/− mice; J.X. Lin (National Heart, Lung and Blood Institute, National Institutes of Health) for discussions and real-time PCR primers; J.F. Zhu, H.H. Xue, H.P. Kim and R. Spolski for discussions and technical assistance; L. Hennighausen for support and discussions; and X. Shirley Liu for the 'model-based analysis of ChiP-seq' algorithm. Supported by the Division of Intramural Research of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Author information
Authors and Affiliations
Contributions
W.L. designed and did research, analyzed data, and wrote the paper; D.E.S. analyzed data and wrote the paper; J.O., Y.C., K.C. and T.Y.R. did research; K.Z. analyzed data; W.J.L. designed research, analyzed data and wrote the paper; and all authors reviewed the paper before submission.
Corresponding author
Supplementary information
Supplementary Text and Figure
Supplementary Figures 1–2, Tables 1–10 and Supplementary Methods (PDF 2647 kb)
Rights and permissions
About this article
Cite this article
Liao, W., Schones, D., Oh, J. et al. Priming for T helper type 2 differentiation by interleukin 2–mediated induction of interleukin 4 receptor α-chain expression. Nat Immunol 9, 1288–1296 (2008). https://doi.org/10.1038/ni.1656
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1656
This article is cited by
-
IL-6 prevents Th2 cell polarization by promoting SOCS3-dependent suppression of IL-2 signaling
Cellular & Molecular Immunology (2023)
-
Strategies to therapeutically modulate cytokine action
Nature Reviews Drug Discovery (2023)
-
Effects of the STAMP-inhibitor asciminib on T cell activation and metabolic fitness compared to tyrosine kinase inhibition by imatinib, dasatinib, and nilotinib
Cancer Immunology, Immunotherapy (2023)
-
The ubiquitin ligase Cul5 regulates CD4+ T cell fate choice and allergic inflammation
Nature Communications (2022)
-
Antimalarial drugs—are they beneficial in rheumatic and viral diseases?—considerations in COVID-19 pandemic
Clinical Rheumatology (2022)