Abstract
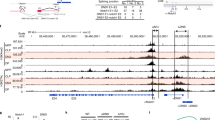
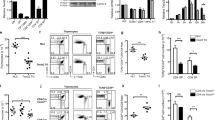
The transcription factor ThPOK is required and sufficient for the generation of CD4+CD8− thymocytes, yet the mechanism by which ThPOK orchestrates differentiation into the CD4+ helper T cell lineage remains unclear. Here we used reporter mice to track the expression of transcription factors in developing thymocytes. Distal promoter–driven expression of the gene encoding the transcription factor Runx3 was restricted to major histocompatibility complex (MHC) class I–selected thymocytes. In ThPOK-deficient mice, such expression was derepressed in MHC class II–selected thymocytes, which contributed to their redirection to the CD8+ T cell lineage. In the absence of both ThPOK and Runx, redirection was prevented and cells potentially belonging to the CD4+ lineage, presumably specified independently of ThPOK, were generated. Our results suggest that MHC class II–selected thymocytes are directed toward the CD4+ lineage independently of ThPOK but require ThPOK to prevent Runx-dependent differentiation toward the CD8+ lineage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kappes, D.J., He, X. & He, X. CD4–CD8 lineage commitment: an inside view. Nat. Immunol. 6, 761–766 (2005).
Zou, Y.R. et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 29, 332–336 (2001).
Sawada, S., Scarborough, J.D., Killeen, N. & Littman, D.R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994).
Kioussis, D. & Ellmeier, W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2, 909–919 (2002).
Laslo, P. et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126, 755–766 (2006).
He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005).
Sun, G. et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6, 373–381 (2005).
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002).
Woolf, E. et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA 100, 7731–7736 (2003).
Egawa, T., Tillman, R.E., Naoe, Y., Taniuchi, I. & Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007).
Sato, T. et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22, 317–328 (2005).
Naoe, Y. et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 204, 1749–1755 (2007).
Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008).
He, X. et al. CD4–CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008).
Levanon, D. et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21, 3454–3463 (2002).
Li, Q.L. et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109, 113–124 (2002).
Grueter, B. et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J. Immunol. 175, 1694–1705 (2005).
Barthlott, T., Kohler, H., Pircher, H. & Eichmann, K. Differentiation of CD4highCD8low coreceptor-skewed thymocytes into mature CD8 single-positive cells independent of MHC class I recognition. Eur. J. Immunol. 27, 2024–2032 (1997).
Chan, S., Correia-Neves, M., Dierich, A., Benoist, C. & Mathis, D. Visualization of CD4/CD8 T cell commitment. J. Exp. Med. 188, 2321–2333 (1998).
Guidos, C.J., Danska, J.S., Fathman, C.G. & Weissman, I.L. T cell receptor-mediated negative selection of autoreactive T lymphocyte precursors occurs after commitment to the CD4 or CD8 lineages. J. Exp. Med. 172, 835–845 (1990).
Kydd, R., Lundberg, K., Vremec, D., Harris, A.W. & Shortman, K. Intermediate steps in thymic positive selection. Generation of CD4−8+ T cells in culture from CD4+8+, CD4int8+, and CD4+8int thymocytes with up-regulated levels of TCR-CD3. J. Immunol. 155, 3806–3814 (1995).
Lucas, B. & Germain, R.N. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity 5, 461–477 (1996).
Lundberg, K., Heath, W., Kontgen, F., Carbone, F.R. & Shortman, K. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4−8+TCRhi thymocytes. J. Exp. Med. 181, 1643–1651 (1995).
Suzuki, H., Punt, J.A., Granger, L.G. & Singer, A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity 2, 413–425 (1995).
Keefe, R., Dave, V., Allman, D., Wiest, D. & Kappes, D.J. Regulation of lineage commitment distinct from positive selection. Science 286, 1149–1153 (1999).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Dave, V.P., Allman, D., Keefe, R., Hardy, R.R. & Kappes, D.J. HD mice: a novel mouse mutant with a specific defect in the generation of CD4+ T cells. Proc. Natl. Acad. Sci. USA 95, 8187–8192 (1998).
Hernandez-Hoyos, G., Anderson, M.K., Wang, C., Rothenberg, E.V. & Alberola-Ila, J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94 (2003).
Pai, S.Y. et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875 (2003).
Waltzer, L., Ferjoux, G., Bataille, L. & Haenlin, M. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 22, 6516–6525 (2003).
Fossett, N., Hyman, K., Gajewski, K., Orkin, S.H. & Schulz, R.A. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc. Natl. Acad. Sci. USA 100, 11451–11456 (2003).
Sarafova, S.D. et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity 23, 75–87 (2005).
Singer, A. & Bosselut, R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83, 91–131 (2004).
Zijlstra, M. et al. Beta 2-microglobulin deficient mice lack CD4−8+ cytolytic T cells. Nature 344, 742–746 (1990).
Grusby, M.J., Johnson, R.S., Papaioannou, V.E. & Glimcher, L.H. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science 253, 1417–1420 (1991).
Lee, P.P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001).
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93, 5860–5865 (1996).
Grogan, J.L. et al. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14, 205–215 (2001).
Acknowledgements
We thank the Rockefeller University Gene Targeting Facility for the generation of ThPOK mutant mice with B6-C2J embryonic stem cells; J.H. Dong, A. Auerbach and W. Ho for microinjection; members of the laboratory for discussion; and J. Huh, M. Chong, A. Collins and S. Schwab for critical reading of the manuscript. Supported by the Leukemia and Lymphoma Society (T.E.) and the Howard Hughes Medical Institute (D.R.L.).
Author information
Authors and Affiliations
Contributions
T.E. did all the experiments; and T.E. and D.R.L. designed the experiments and wrote the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Table 1 (PDF 5056 kb)
Rights and permissions
About this article
Cite this article
Egawa, T., Littman, D. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol 9, 1131–1139 (2008). https://doi.org/10.1038/ni.1652
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1652
This article is cited by
-
Markers and makers of NKT17 cells
Experimental & Molecular Medicine (2023)
-
Runx3d controls the abundance and functional differentiation of CD4+CD8αα+ intraepithelial T cells
Cell Death Discovery (2023)
-
Unique properties of tissue-resident memory T cells in the lungs: implications for COVID-19 and other respiratory diseases
Nature Reviews Immunology (2023)
-
Integrative scATAC-seq and scRNA-seq analyses map thymic iNKT cell development and identify Cbfβ for its commitment
Cell Discovery (2023)
-
Reversal of the T cell immune system reveals the molecular basis for T cell lineage fate determination in the thymus
Nature Immunology (2022)