Abstract
Robust production of type I interferon (IFN-α/β) in plasmacytoid dendritic cells (pDCs) is crucial for antiviral immunity. Here we show involvement of the mammalian target of rapamycin (mTOR) pathway in regulating interferon production by pDCs. Inhibition of mTOR or its 'downstream' mediators, the p70 ribosomal S6 protein kinases p70S6K1 and p70S6K2, during pDC activation by Toll-like receptor 9 (TLR9) blocked the interaction of TLR9 with the adaptor MyD88 and subsequent activation of the interferon-regulatory factor IRF7, which resulted in impaired IFN-α/β production. Microarray analysis confirmed that inhibition of mTOR by the immunosuppressive drug rapamycin suppressed antiviral and anti-inflammatory gene expression. Consistent with this, targeting rapamycin-encapsulated microparticles to antigen-presenting cells in vivo resulted in less IFN-α/β production in response to CpG DNA or the yellow fever vaccine virus strain 17D. Thus, mTOR signaling is crucial in TLR-mediated IFN-α/β responses by pDCs.
Similar content being viewed by others
Main
Plasmacytoid dendritic cells (pDCs) are specialized immune cells able to rapidly produce large amounts of type I interferon for antiviral innate immunity1,2,3. Induction of the antiviral innate immune response depends on recognition of viral components by Toll-like receptors (TLRs)4. The pDCs 'preferentially' express virus-recognizing TLR7 and TLR9, which sense viral single-stranded RNA and unmethylated CpG-rich DNA, respectively5. 'Downstream' of TLR7 and TLR9 engagement, interferon production by pDCs depends absolutely on the adaptor protein MyD88–interferon-regulatory protein IRF7 signaling pathway6,7. Data have shown that phosphatidylinositol-3-OH kinase (PI(3)K)–Akt kinase activity influences TLR-induced cytokine production in myeloid DCs8. The kinase mTOR (mammalian target of rapamycin) is a crucial 'downstream' mediator of PI(3)K-Akt signaling and a key regulator of cell growth and proliferation9,10,11. Increasing evidence suggests that mTOR integrates signals from nutrients, energy status and growth factors to regulate many processes, including autophagy, ribosome biogenesis and metabolism9,10,11. It has been found that mTOR also is important in the survival and maturation of myeloid DCs12, which suggests there is crosstalk between TLR signaling and the PI(3)K-Akt-mTOR pathway. However, it is unclear whether mTOR influences the function of pDCs.
Here we show that the rapamycin-sensitive PI(3)K-mTOR–p70 ribosomal S6 protein kinase (p70S6K) pathway is crucial for TLR-mediated induction of type I interferon in pDCs. Inhibition of mTOR or p70S6K by various approaches resulted in much less TLR-mediated production of interferon-α (IFN-α) by pDCs. The mechanism of this inhibition seemed to involve disruption of the TLR9-MyD88 complex and subsequent impairment of phosphorylation and nuclear translocation of IRF7. A physiological function for the mTOR pathway in regulating IFN-α production was emphasized by the finding that mTOR inhibition in vivo resulted in less IFN-α in serum and in pDCs in response to stimulation with a TLR ligand or viral vaccine, which ultimately led to impaired immune responses.
Results
Rapamycin inhibits TLR-mediated IFN-α production by pDCs
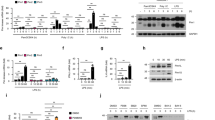
To assess the involvement of mTOR in pDC function, we used the highly specific mTOR inhibitor rapamycin13. We stimulated purified human and mouse pDCs with the TLR9 ligand A-D-type CpG oligodeoxynucleotide (CpG-A; ODN 2336)14 in the presence of rapamycin or with vehicle alone. Rapamycin, even at low nanomolar concentrations, substantially suppressed CpG-A-induced IFN-α production by both mouse and human pDCs (Fig. 1a). In addition to IFN-α, activated pDCs secrete IFN-β and moderate amounts of other proinflammatory cytokines, including tumor necrosis factor (TNF) and interleukin 6 (IL-6). Rapamycin also significantly inhibited pDC secretion of IFN-β, TNF and IL-6 (Fig. 1b and Supplementary Fig. 1 online). In contrast, production of CXCL10 (IP-10) was induced by CpG but was not suppressed by rapamycin. The inhibitory effect of rapamycin on IFN-α production was not limited to TLR9 stimulation, as rapamycin also impaired IFN-α production induced by the TLR7 ligand loxoribine and several viruses, including the yellow fever vaccine strain 17D (YF-17D) and vesicular stomatitis virus (Fig. 1c). To exclude the possibility that these effects were caused simply by a decrease in pDC numbers through apoptosis, we examined the apoptotic sensitivity of pDCs to mTOR inhibition. Rapamycin, even at high concentrations, did not induce apoptosis of pDCs, as shown by labeling with annexin V and propidium iodide (Fig. 1d).
(a) ELISA of IFN-α in supernatants of isolated pDCs pretreated for 3 h with various doses of rapamycin (Rap) and then stimulated with CpG-A (10 μg/ml; left) or treated with vehicle (Veh) or rapamycin and then stimulated with various doses of CpG-A (right). (b) ELISA of IFN-β, TNF, IL-6 and CXCL10 in supernatants of purified mouse pDCs pretreated with various doses of rapamycin and then stimulated for 24 h with CpG-A (10 μg/ml). (c) ELISA of IFN-α in supernatants of purified spleen pDCs cultured for 24 h with CpG-A (10 μg/ml), loxoribine (Loxo; 1 mM), YF-17D or vesicular stomatitis virus (VSV; all viruses at a multiplicity of infection of 1) after pretreatment with rapamycin (Rap) or vehicle. (d) Apoptotic death of isolated mouse pDCs pretreated with vehicle or 20 nM or 500 nM rapamycin, assessed by staining with annexin V and propidium iodide. Numbers in quadrants indicate percent cells in each. (e) Proliferation of and cytokine induction in naive CD4+CD62L+ OT-II T cells (1 × 105 cells/well) cultured with pDCs (5 × 104) pretreated with ovalbumin peptide (10 μg/ml) and CpG-A (10 μg/ml) plus vehicle or rapamycin (100 nM for 3 h), assessed after 3 d by incorporation of [3H]thymidine (top left; proliferation) and ELISA of IFN-γ, IL-4, IL-10, IL-13 and IL-17. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. Data are representative of at least three independent experiments (error bars, s.d.).
To determine the effect of rapamycin on other DC subsets, we analyzed mouse bone marrow–derived DCs and human monocyte-derived DCs as described above. In both types of DCs, there was substantial diminution of IL-10, IL-6 and TNF production when cells were treated with rapamycin before begin activated with lipopolysaccharide (LPS; Supplementary Fig. 2 online). In contrast, IL-12p40 production in mouse bone marrow–derived DCs and IL-12p70 production in human monocyte-derived DCs were not affected by rapamycin pretreatment (Supplementary Fig. 2). In addition to rapamycin, cyclosporin A (CsA) and FK506 (tacrolimus) are two other commonly used immunosuppressive drugs. FK506 has structural similarity to rapamycin and even forms a complex with the same FK506 binding proteins15. Both CsA and FK506 bind to and inhibit calcineurin in a calcium-dependent signaling pathway16. CsA and FK506 had different effects on cytokine production by bone marrow– and monocyte-derived DCs than did rapamycin. CsA and FK506 considerably suppressed IL-12p70 production but had no effect on the production of IL-12p40, IL-10, IL-6 and TNF by LPS-activated bone marrow–derived DCs and monocyte-derived DCs (Supplementary Fig. 2). Collectively, these data show that inhibition of mTOR suppresses IFN-α/β production by pDCs after microbial stimulation.
Given that rapamycin suppressed the TLR-mediated innate responses of pDCs, we determined whether rapamycin-treated pDCs were compromised in their ability to stimulate T cell responses. We used an in vitro antigen presentation assay in which we cultured pDCs with ovalbumin, CpG-A and OT-II CD4+ T cells, which express a transgenic T cell receptor specific for ovalbumin peptide presented by major histocompatibility complex class II molecules. After 3 d of culture, we assessed the proliferation of OT-II T cells through their incorporation of tritiated thymidine and measured the induction of IFN-γ, IL-4, IL-10, IL-13 and IL-17 by enzyme-linked immunosorbent assay (ELISA). The pDCs pulsed with CpG-A and ovalbumin induced robust OT-II T-cell proliferation; in contrast, rapamycin-pretreated pDCs showed a reduced immune-stimulatory capacity (Fig. 1e). We next assessed cytokine production by the OT-II cell cultures described above. In cultures from vehicle-treated pDCs, we detected large amounts of IFN-γ, IL-17, IL-10, IL-4 and IL-13; in contrast, rapamycin-pretreated pDCs had much lower expression of IFN-γ, IL-17 and IL-10 and had slightly lower expression of the T helper type 2 (TH2) cytokines IL-4 and IL-13. These data indicate that rapamycin-treated pDCs are impaired in their ability to stimulate antigen-specific CD4+ T cell activation in vitro, consistent with a published study17.
TLR stimulation of pDCs activates the mTOR pathway
To assess the mechanism by which rapamycin inhibits TLR-mediated interferon production by pDCs, we examined the activation of mTOR by detecting its phosphorylated form with a specific antibody to phosphorylated mTOR. CpG-A strongly induced phosphorylation of mTOR in the RAW264.7 mouse macrophage cell line within 15 min and for at least 1 h; rapamycin pretreatment inhibited the effect of CpG-A at all time points tested (Fig. 2a). Unexpectedly, in pDCs, we found constitutive phosphorylation of mTOR (at the serine at position 2448 (Ser2448)) even in the absence of stimulation with CpG-A, although pretreatment of pDCs with rapamycin inhibited mTOR phosphorylation (Fig. 2b).
(a) Immunoblot analysis of phosphorylated (phospho-) and total mTOR in lysates of RAW cells (4 × 106) transiently transfected for 40 h with cyan fluorescent protein–tagged MyD88 and IRF7-YFP, treated with vehicle (–) or rapamycin (+) and then stimulated for 0–60 min with CpG-A–DOTAP. Results are representative of two independent experiments. (b) Immunoblot analysis of phosphorylated and total mTOR, p70S6K, 4E-BP1 and Akt in lysates of purified pDCs (1 × 106) stimulated for 15 min with CpG-A in the presence (Rap) or absence (Medium) of rapamycin. Results are representative of three independent experiments.
Phosphorylation of mTOR leads to more phosphorylation and inactivation of the repressor of mRNA translation 4E-BP1 (eukaryotic initiation factor 4E–binding protein). Phosphorylation of mTOR also leads to activation of p70S6K, which enhances protein translation9,10,11,12,18,19,20,21. It has been shown that mTOR, p70S6K and 4E-BP1 are all 'downstream' phosphorylation targets of the kinase Akt22. As with mTOR, p70S6K and Akt were constitutively phosphorylated in freshly isolated pDCs (Fig. 2b). Stimulation of pDCs with CpG-A resulted in a slight, yet rapid, enhancement of p70S6K phosphorylation at 5 min, which decreased to constitutive baseline by 15 min (data not shown), and a more substantial enhancement of 4E-BP1 phosphorylation. Pretreatment of pDCs with rapamycin inhibited the phosphorylation of p70S6K at Thr389. Initially, rapamycin did not seem to affect the phosphorylation of 4E-BP1 at Thr37, Thr46 or Thr70 much, although it did diminish phosphorylation at Ser65 (Fig. 2b); however, exposure to rapamycin for 6 h or longer did result in inhibition of phosphorylation at Thr70 (data not shown). The total amounts of mTOR, p70S6K and 4E-BP1 protein were not altered much by rapamycin.
TLR induction of IFN-α requires the mTOR pathway
We next determined the functional consequences of disrupting the mTOR pathway in pDCs. Knockdown of mTOR in pDCs by small interfering RNA (siRNA) suppressed IFN-α production after CpG-A stimulation (Fig. 3a), consistent with the inhibitory effect of rapamycin on IFN-α production. Pretreatment of cells with LY294002, a specific inhibitor of PI(3)K (an 'upstream' mediator of mTOR21), also considerably suppressed CpG-A-induced IFN-α secretion in pDCs in a dose-dependent way (Fig. 3b). This indicated that the PI(3)K-mTOR pathway is crucial for CpG-A-induced production of IFN-α in pDCs.
(a) ELISA of IFN-α in supernatants (above) and immunoblot analysis of mTOR in lysates (below) of purified mouse pDCs (5 × 105) transfected for 5 h with control or mTOR-specific siRNA and then stimulated for 24 h with CpG-A. Below, β-actin serves as a loading control. (b) ELISA of IFN-α in supernatants of pDCs treated with rapamycin or the PI(3)K inhibitor LY294002 (1–25 μM), then stimulated with CpG-A and assessed 24 h later. (c) ELISA of IFN-α in supernatants (above) and immunoblot analysis of p70S6K in lysates (below) of mouse pDCs transfected for 5 h with siRNA pools specific for p70S6K1 (S6K1), p70S6K2 (S6K2) or both (S6K1,2), then stimulated for 24 h with CpG-A. *, P < 0.05. (d) ELISA of IFN-α in supernatants of pDCs isolated from wild-type (WT) and Rps6k1−/− Rps6k2−/− double-knockout (S6K1,2-KO) mouse spleens and stimulated in vitro with CpG-A. Data are representative of three independent experiments (a–c) or two independent experiments with at least two mice per group per experiment (d); error bars, s.e.m. of replicate wells.
Deletion of 4E-BP1 and 4E-BP2 has been shown to enhance the production of type I interferon in pDCs through translational control of IRF7 mRNA23. However, it is not clear whether p70S6K also regulates IFN-α production in pDCs. We therefore evaluated the function of p70S6K in this process. We transfected purified pDCs with siRNA pools specific for only p70S6K1 or p70S6K2 mRNA or with siRNA pools specific for both. We confirmed the efficacy of the siRNA in 'knocking down' gene expression by measuring protein (Fig. 3c). Downregulation of p70S6K1 (Rps6kb1) or p70S6K2 (Rps6kb2) by siRNA resulted in less IFN-α secretion by mouse pDCs (Fig. 3c); knockdown of both kinases resulted in even less secretion of IFN-α (Fig. 3c). Consistent with those findings, there was less IFN-α secretion by pDCs isolated from Rps6k1−/− Rps6k2−/− double-knockout mice (Fig. 3d). However, the spleens of the double-knockout mice did not have fewer pDCs (Supplementary Fig. 3 online). These results collectively show that CpG-A-induced IFN-α production in pDCs is mediated by the PI(3)K-mTOR-p70S6K signaling pathway.
Blocking TLR9-MyD88 interaction and IRF7 phosphorylation
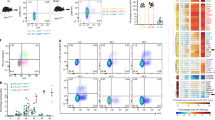
To further understand the mechanism of action of rapamycin in its inhibition of IFN-α in pDCs, we first assessed by flow cytometry the intracellular expression of TLR9 in rapamycin-treated pDCs. TLR9 was constitutively expressed in pDCs, and there was no substantial difference in TLR9 expression by CpG-A-activated pDCs and resting pDCs (Fig. 4a). Rapamycin did not alter pDC TLR9 expression. The mechanism by which rapamycin inhibits IFN-α production therefore probably does not involve less expression of TLR9. Stimulation of DCs with CpG causes both TLR9 and MyD88 to be rapidly redistributed toward sites of CpG accumulation24. To address whether rapamycin treatment affected TLR9 redistribution and its subsequent binding to CpG, we exposed RAW cells expressing yellow fluorescent protein (YFP)-tagged TLR9 to indodicarbocyanine-tagged CpG-A and examined the localization of TLR9 and CpG-A by confocal microscopy. Rapamycin did not seem to affect the association of TLR9 with CpG-A (Fig. 4b).
(a) Flow cytometry of isolated spleen pDCs stimulated with CpG-A with or without rapamycin pretreatment, then stained for intracellular TLR9. (b) Deconvolution microscopy of RAW cells expressing YFP-tagged TLR9 (green), pretreated with vehicle (left) or rapamycin (right) and then incubated for 90 min with indodicarbocyanine-tagged CpG-A–DOTAP (red; 5 μg/ml). Two images were obtained for each condition. Data are representative of two independent experiments.
TLR9 signaling results in MyD88-dependent activation of IRF7 and the adaptor protein TRAF6, which together mediate IFN-α production7,25. We further evaluated the effect of rapamycin on this signaling pathway in RAW cells, which express type I interferons. We stimulated RAW cells coexpressing YFP-tagged TLR9 and hemagglutinin-tagged MyD88 with the cationic lipid DOTAP (CpG-A–DOTAP)7 to activate the TLR9-MyD88-IRF7 signaling pathway. We then lysed the cells, immunoprecipitated equal amounts of proteins with antibody to hemagglutinin (anti-hemagglutinin) and used immunoblot analysis to detect TLR9. TLR9 immunoprecipitated together with MyD88 after stimulation with CpG-A–DOTAP; this interaction was prevented by rapamycin (Fig. 5a). We further verified the interaction between TLR9 and MyD88 using various expression constructs and by converse experiments in which we immunoprecipitated TLR9-MyD88 complexes with anti-TLR9 and then detected them with anti-MyD88 (Supplementary Fig. 4 online and data not shown). In each of these cases, rapamycin inhibited the formation of TLR9-MyD88 complexes; consistent with this, siRNA knockdown of p70S6K1 and p70S6K2 also impaired TLR9-MyD88 interaction (Supplementary Fig. 4).
(a) Immunoassay of RAW cells (4 × 106) transfected with hemagglutinin-tagged MyD88 and YFP-tagged TLR9 and then, 36 h later, stimulated for 90 min with CpG-A–DOTAP; cell extracts were immunoprecipitated (IP) with anti-hemagglutinin and analyzed by immunoblot (IB) for coprecipitated TLR9. MyD88 serves as a loading control. (b) Flow cytometry of human pDCs pretreated with rapamycin (left) or transfected with control (con) siRNA or siRNA specific for p70S6K1 and p70S6K2 (S6K), then stimulated for 45 min with CpG-A; cells were fixed, made permeable and stained with phycoerythrin-conjugated mouse antibody to IRF7 phosphorylated at Ser477 and Ser479 (phosphor-IRF7) or control isotype antibody (Control Ab; phycoerythrin-conjugated mouse immunoglobulin G1). (c) Confocal microscopy of purified mouse pDCs stimulated for 12 h with CpG-A in the presence of rapamycin or vehicle, then fixed with formaldehyde, made permeable with saponin, blocked with 20% (vol/vol) goat serum, stained with anti-IRF7 (green) and mounted with ProLong Gold antifade reagent with DAPI (blue). Scale bar, 2 μm. (d) Luciferase assay of HEK293 cells transiently transfected with 0 ng (–), 40 ng or 200 ng of constitutively active IRF7 (IRF7-D477,479) together with luciferase-tagged IFN-β (plasmid p125-Luc; 50 ng), renilla luciferase (0.5 ng), wild-type IRF7 (3 ng), MyD88 (20 ng) and TLR9 (50 ng), cultured for 24 h, then pretreated for 3 h with 100 nM rapamycin and then stimulated overnight with CpG-A (10 μg/ml). Empty vector, vector without IRF7. Data are representative of three (a,d) or two (b,c) independent experiments (error bars (d), s.d.).
IRF7 regulates type I interferon–dependent immune responses6. Central to the activation of IRF7 is its phosphorylation, which leads to the formation of homodimers of IRF7 and nuclear accumulation of dimers that are able to bind DNA and transactivate interferon target genes26,27. To study the effect of rapamycin on the function of IRF7, we pretreated purified human pDCs for 3 h with rapamycin and then with CpG-A. We then detected phosphorylation of IRF7 in pDCs with an antibody to IRF7 phosphorylated at Ser477 and Ser479. In the absence of rapamycin, CpG-A activation of pDCs led to phosphorylation of IRF7; phosphorylation was inhibited by rapamycin treatment, although total IRF7 protein expression was unaffected (Fig. 5b and Supplementary Fig. 5 online). Similarly, knockdown of p70S6K1 and p70S6K2 with specific siRNA pools resulted in less IRF7 phosphorylation (Fig. 5b). Moreover, activation with CpG-A for 12 h led to IRF7 localization mainly in the nucleus in most isolated pDCs; this nuclear translocation of IRF7 was inhibited by pretreatment with rapamycin (Fig. 5c). We also overexpressed a constitutively active form of IRF7 in human embryonic kidney (HEK293) cells before CpG-A stimulation and found that this 'rescued' the defect in interferon production after mTOR inhibition (Fig. 5d). These results indicate that rapamycin inhibits the activation and nuclear translocation of IRF7 in pDCs.
We next used microarray to analyze gene expression in sorted mouse pDCs to obtain further insights into the mechanism by which rapamycin inhibited IFN-α induction in pDCs. After CpG-A stimulation for 4 or 12 h, pDCs had higher expression of genes encoding several antiviral proteins (such as IFN-α4, IFN-β1 and the interferon-inducible proteins IFI44 and IFI202B) and inflammatory molecules, including the complement proteins C5AR1 and C3, as well as several chemokines (CCL2, CCL7, CCL8 and CCL12) and chemokine receptors (CCR3). In the presence of rapamycin, expression of the genes encoding these proteins was lower, whereas the expression of genes encoding other proteins, such as CXCL10, CD86, IL-12B and IL-8RB, was either not lower or was actually higher in the presence of rapamycin (Supplementary Fig. 6 online).
In vivo targeting of rapamycin
To determine whether rapamycin inhibited type I interferon production in vivo, we injected rapamycin into C57BL/6 mice and, after 3 d, injected CpG-A or live attenuated YF-17D. YF-17D, one of the most effective vaccines28, induces immune signaling through many TLRs, including TLR2, TLR7, TLR8 and TLR9 (ref. 29). We used ELISA to measure the kinetics of serum production of IFN-α and IFN-α produced by pDCs isolated and cultured ex vivo. Injection of CpG-A induced robust IFN-α production in pDCs 12 h after stimulation, which decreased slowly over 48 h (Supplementary Fig. 7 online). YF-17D-activated secretion of IFN-α had similar kinetics, and pretreatment with rapamycin resulted in significantly less IFN-α (Fig. 6a). In contrast to the robust CpG-A-mediated induction of IFN-α in pDCs, we detected no IFN-α in the serum of CpG-A-induced mice at any time points tested (Supplementary Fig. 7). However, we did detect robust serum IFN-α secretion in response to YF-17D, which peaked at 12 h and decreased to baseline at 24 h; the administration of rapamycin before YF-17D challenge inhibited serum IFN-α production (Fig. 6a). Knockdown of p70S6K1 and p70S6K2 in pDCs inhibited the production of IFN-α in response to YF-17D in vitro (Supplementary Fig. 8 online), which supported our data obtained with rapamycin (Fig. 1c). These results indicate that rapamycin potently inhibits the secretion of IFN-α in pDCs in vivo in response to CpG-A and YF-17D.
(a) ELISA of IFN-α production by pDCs from C57BL/6 mice treated daily for 3 d with soluble rapamycin (1.5 mg per kg body weight per day) or rapamycin encapsulated in PLGA microparticles (2 mg per mouse per day), then vaccinated subcutaneously with YF-17D on day 4; blood and spleens were obtained at various times after injection for analysis of serum (right) and pDCs enriched from spleens with microbeads coated with anti–mouse PDCA1 (left). *P < 0.05; **P < 0.001; ***P < 0.0001. Data are representative of at least three independent experiments (error bars , s.d.). (b) Flow cytometry of ovalbumin (OVA) in cells from C57BL/6 mice left untreated (top row) or subcutaneously injected with Alexa Fluor 488–labeled ovalbumin encapsulated in PLGA microparticles (bottom row); draining lymph nodes were isolated 24 h later and treated for 30 min at 37 °C with collagenase type IV, then isolated cells were stained for cell surface lineage markers to identify cell type (above plots). Numbers below outlined areas indicate percent cells containing OVA. cDCs, conventional DCs. Data are representative of three independent experiments.
Because soluble rapamycin targets all cells in vivo and therefore would not have a specific inhibitory effect on DCs16, we encapsulated rapamycin in poly(lactic-co-glycolic) acid (PLGA) microparticles30 and injected these into mice intraperitoneally to specifically enhance uptake of rapamycin by antigen-presenting cells (APCs). Published work has shown that PLGA microparticles can enhance the delivery of encapsulated molecules to APCs30, and our initial results confirmed that (Fig. 6b). Additional experiments showed that pDCs from mice treated with PLGA-encapsulated rapamycin secreted much less IFN-α both after infection with YF-17D and after systemic administration of CpG-A (Fig. 6a and Supplementary Fig. 7).
To investigate the effects of rapamycin delivery to APCs on T cell responses in vivo, we immunized mice with YF-17D and analyzed the antigen-specific CD8+ T cell response in the draining lymph nodes. We prepared single-cell suspensions of lymph nodes 5 d later and restimulated for 72 h in vitro with a combination of yellow fever CD8+ T cell epitope peptides from NS3 and E-1 (ref. 29). We then evaluated the induction of IFN-γ, IL-17, IL-4, IL-13 and IL-10 (TH1, TH2 and IL-17-producing T helper cell cytokines) in culture by ELISA and intracellular flow cytometry. YF-17D induced IFN-γ production as measured in total cell culture supernatants and in CD8+ T cells (Fig. 7). YF-17D also induced the production of TH2 cytokines IL-4, IL-13 and IL-10, consistent with a published report that YF-17D induces a mixed TH1 and TH2 T cell response in vivo29. IL-17 production was also induced by in vivo YF-17D stimulation. In vivo administration of PLGA-encapsulated rapamycin before virus challenge inhibited TH1, TH2 and L-17-producing T helper cell cytokines. Collectively, these findings show that mTOR inhibition in APCs in vivo can suppress the adaptive immune response to a vaccine.
IFN-γ production (top right) and cytokine expression (top left and bottom) by CD8+ T cells enriched from draining lymph nodes of C57BL/6 mice left untreated (–) or treated daily for 3 d with vehicle or rapamycin microparticles (2 mg per mouse per day) and then vaccinated subcutaneously on day 4 with YF-17D; 5 d later, single-cell suspensions were cultured for an additional 3 d in vitro in medium alone (left bars) or with a CD8+ T cell epitope from YF-17D (middle and right bars). IFN-γ was assessed by intracellular cytokine staining and flow cytometry; numbers above outlined areas indicate percent CD8+IFN-γ+ cells. IFN-γ, IL-4, IL-13, IL-10 and IL-17 in cell supernatants were measured by ELISA. *, P < 0.05. Data are representative of three independent experiments (error bars, s.d.).
Discussion
It is well established that engagement of TLRs by many viruses stimulates robust production of type I interferon in pDCs, but the molecular mechanisms that regulate this important antiviral response are not fully understood. Given the crucial function of type I interferons in innate and adaptive immunity, the interferon response must be under tight control to prevent pathogenic responses. Many regulatory mechanisms involving surface receptors (BDCA2, ILT7, Siglec-H, immunoglobulin E receptor and NKp44), intracellular and exogenous factors (IRF4), and virus-encoded molecules have been shown to modulate the interferon responses of pDCs1,2,3. Osteopontin is reported to be essential for TLR9-dependent interferon production by pDCs31. Here we have shown that mTOR has a crucial function in the TLR-mediated induction of IFN-α in pDCs, in vitro and in vivo. Using several independent methods to investigate whether the mTOR signaling pathway regulates type I interferon activity, we found that inhibition of mTOR-p70S6K resulted in inhibition of IRF7 phosphorylation and nuclear localization, which impaired the production of type I interferon.
Several aspects of our study warrant further comment. The first relates to the mechanism by which rapamycin inhibits TLR-mediated IFN-α production in pDCs. Our data indicated that knockdown of mTOR or its downstream mediator p70S6K resulted in disruption of the MyD88-TLR9 interaction and nuclear localization of IRF7. Activation of mTOR enhances translation initiation in part by phosphorylation of its main targets, 4E-BP1, 4E-BP2, p70S6K1 and p70S6K2 (refs. 10,11,12,18). Both 4E-BP1 and 4E-BP2 are translational repressors, and their phosphorylation by mTOR abrogates their ability to bind the initiation factor eIF4E and inhibit protein synthesis32. A study has shown that inhibiting expression of 4E-BP1 and 4E-BP2 enhances IRF7 translation and increases virus-stimulated interferon synthesis23. Our study has indicated that in addition to the 4E-BP-dependent translational regulation of IRF7, activation of p70S6K leads to enhanced IRF7 phosphorylation and nuclear translocation. Together, the previous study23 and our study here indicate that mTOR-mediated phosphorylation of 4E-BP1, 4E-BP2, p70S6K1 and p70S6K2 promotes IFN-α production through IRF7. Our results are consistent with a study reporting a crucial function for PI(3)K in nuclear translocation of IRF7 and production of type I interferon by human pDCs in response to TLR stimulation33, although the involvement of the 'downstream' mTOR pathway was not reported in that study.
The effect of rapamycin in blocking the association of TLR9 and MyD88 raises the issue of whether there is direct physical interaction between the mTOR complex and TLR9-MyD88. We were unable to obtain evidence of such an interaction by immunoprecipitation studies (data not shown). In addition, because TLR9-MyD88 interaction is a proximal event, it seems likely that rapamycin treatment would also inhibit other TLR9-mediated pathways, such as the transcription factor NF-κB pathway. Rapamycin treatment did indeed inhibit TLR9-mediated NF-κB signaling (data not shown), consistent with the lower production of proinflammatory cytokines such as TNF and IL-6. However, the lack of any effect on expression of CXCL10 or proteins such as CD86 may reflect incomplete blockage of the TLR9-MyD88 association by rapamycin.
Another issue is the extent to which the effects of rapamycin are specific to the TLR7-TLR9-MyD88-IRF7 axis in pDCs. To clarify the TLR specificity of mTOR inhibition, we examined IFN-α/β production by mouse splenic CD11c+ DCs in response to various TLR ligands, as pDCs express only a limited set of TLRs. Rapamycin blocked IFN-α secretion in response to stimulation by ligands for TLR4, TLR7 and TLR9, but not in response to polyinosinic-polycytidylic acid (data not shown). TLR9 and TLR7 are thought to mediate all their effects through MyD88; in the case of LPS, the adaptor protein TRIF is believed to mediate the induction of IFN-β34. However, it is not clear whether TRIF is directly involved in the induction of IFN-α. In fact, it has been reported25 that IFN-α is induced by LPS indirectly: TLR4 activation induces IFN-β through TRIF-IRF3, and this secreted IFN-β induces IRF7 activation, which then induces IFN-α. The effects of rapamycin on the induction of IFN-α in response to LPS can therefore be explained by the finding that knockdown of p70S6K1 and p70S6K2 inhibited IRF7 activation in our studies. In contrast, there was no inhibition of IFN-α production in response to stimulation with polyinosinic-polycytidylic acid (data not shown), which is known to induce type I interferons (IFN-α and IFN-β) through cytosolic helicase and RNA sensor MDA5–dependent signaling with the signaling adaptor IPS-1 (ref. 35). We also noted that rapamycin inhibited IFN-α production in pDCs in response to modified vaccinia virus Ankara (data not shown). The mechanism of induction of IFN-α by this virus is thought to rely on both TLR-independent and TLR-dependent pathways36. Because this virus stimulates many receptors, including TLRs and non-TLRs, it is possible that mTOR also affects signaling through these non-TLRs.
It is notable that we found Akt, mTOR and p70S6K, and, to a lesser extent, 4E-BP1, to be constitutively activated in pDCs in the steady state. One possible explanation for this is that pDCs are very sensitive to various stresses, including 'danger signals' from pathogen products37 and changes in temperature and nutrients. In fact, the process of isolating DCs can cause them to become nonspecifically activated, as shown by the spontaneous upregulation of costimulatory molecules in cell culture in the absence of any exogenous stimulation38. Given the central function of mTOR in regulating cellular responses to fundamental changes in nutrients, growth factors and energy status, it is possible that the stresses involved in their isolation-induced activation of the mTOR pathway39. Immunoblot analysis showed that 'untouched' pDCs, isolated by depletion of other cell types from mouse spleen, had constitutively active mTOR (data not shown). Thus, constitutively phosphorylated mTOR in pDCs may represent a unique adaptation of these cells to facilitate rapid deployment of the type I interferon response against viral infection. In this context, it is notable that other such adaptations to facilitate rapid induction of type I interferons have been reported in pDCs. For example, the interferon-inducing TLR9 ligand CpG-A is retained with the MyD88-IRF7 complex for long periods in the endosomal vesicles of pDCs7. In summary, our data show a function for mTOR in regulating TLR-mediated activation of pDCs. Elements of the mTOR signaling pathway may thus represent potential therapeutic targets in the control of viral immunity and type I interferon–mediated diseases such as lupus40.
Methods
Mice and reagents.
Rps6k1−/− Rps6k2−/− double-knockout mice were provided by S.C. Kozma and N. Sonenberg. C57BL/6 mice were from The Jackson Laboratory. Mice were maintained in specific pathogen–free conditions in the Emory Vaccine Center vivarium. All animal protocols were reviewed and approved by the Institute Animal Care and Use Committee of Emory University. CsA, FK506 (tacrolimus) and rapamycin (sirolimus) were from Sigma. CpG-A (ODN 2336) DNA was from Coley Pharmaceutical indodicarbocyanine-labeled CpG-A was from Sigma Genosys. The PLGA RG502H was from Boehringer Ingelheim Chemicals. Polyvinyl alcohol was from Aldrich Chemicals. Control ON-TARGETplus siCONTROL nontargeting pool and ON-TARGETplus SMART pool for mouse Rps6kb1 and Rps6kb2 were from Dharmacon. The siRNA specific for mouse mTov (FRAP) was from Santa Cruz. Cyan fluorescent protein–tagged MyD88 and YFP-tagged IRF7 plasmids were provided by T. Taniguchi. YFP-tagged TLR9 plasmid (Addgene plasmid 13642) was provided by D. Golenbock. Green fluorescent protein (GFP)-tagged TLR9 was provided by D. Segal. Hemagglutinin-tagged MyD88 plasmid (Addgene plasmid 12287) was provided by B. Beutler. Constitutively active IRF7 (D477,479) was provided by J. Hiscott.
Microbial stimuli.
YF-17D subpassaged from YF-VAX (Aventis Pasteur) in SW-480 cells was provided by R. Ahmed. Modified vaccinia virus Ankara was provided by R.R. Amara. Highly purified Escherichia coli LPS (strain 25922) was provided by T. Van Dyke. Loxoribine (tlrl-lox), imiquimod (R837; tlrl-imq) and polyinosinic-polycytidylic acid (tlrl-pic) were from InvivoGen.
Preparation of DCs.
For the preparation of splenic pDCs, spleens were digested with collagenase type IV (Worthington Biomedical), and samples were enriched for CD45RA+CD11cint pDCs by the use of microbeads coated with anti–mouse PDCA1 (Miltenyi Biotec). For some studies, enriched cells were further sorted with a FACSAria (BD Biosciences). Human pDCs were isolated from peripheral blood mononuclear cells with a pDC isolation kit (Miltenyi Biotec). For the preparation of mouse bone marrow–derived DCs and human monocyte-derived DCs, mouse bone marrow cells or human monocytes were cultured for 6 d with granulocyte-macrophage colony-stimulating factor (10 ng/ml) and IL-4 (PeproTech). For measurement of interferon production, cells were seeded into 96-well plates (2 × 105 cells per well) and stimulated for 24–40 h with various reagents. Interferon in cell supernatants was measured by ELISA (PBL Biomedical Laboratories).
Transfection experiments.
The mouse macrophage cell line RAW264.7 (obtained from S. Zughaier) was transfected with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). DharmaFECT transfection reagent (Dharmacon) was used for transfection of siRNA (100 nM).
T cell assays.
For in vitro stimulation, purified splenic pDCs (5 × 104) were pretreated for 3 h with rapamycin (100 nM) and washed three times. Cells were then cultured with naive CD4+CD62L+ OT-II T cells (1 × 105), CpG-A (10 μg/ml) and ovalbumin (ISQVHAAHAEINEAGR; 10 μg/ml) in 200 μl RPMI complete medium in 96-well round-bottomed plates. After 72 h, supernatants were collected for cytokine measurement and T cell proliferation was assessed as [3H]thymidine incorporation. For analysis of yellow fever–specific T cell responses, isolated lymph node cells from mice injected with YF-17D were plated at a density of 5 × 105 cells per well in 96-well plates and were cultured in complete DMEM with 10% (vol/vol) FBS alone or with a combination of the yellow fever CD8+ T cell epitope peptides NS3 (VIDAMCHATLTYRML) and E-1 (AHCIGITDRDFIEGV). On the third day, supernatants were collected for cytokine measurement. For intracellular cytokine analysis, lymph node cells were stimulated for 6 h with yellow fever peptides in the presence of GolgiStop (PharMingen) and were stained with peridinin chlorophyll protein–conjugated anti-CD8 (53–6.7; Clontech). Cells were made permeable with Cytofix/Cytoperm and were stained with phycoerythrin-conjugated anti-IFN-γ (XMG1.2; Clontech).
Immunoassays.
Mouse spleen pDCs were enriched with microbeads coated with anti–mouse PDCA-1. The purity of each population was routinely between 80% and 85%, as determined by flow cytometry. Isolated pDCs (1 × 106) were pretreated for 3 h with rapamycin and were stimulated for 15 min with CpG-A. Cells were lysed with 100 μl M-PER mammalian protein extraction reagent (Pierce) containing Halt protease inhibitor, EDTA and phosphatase inhibitor (Pierce). Equal amounts of protein were separated by SDS-PAGE and were transferred onto polyvinylidene fluoride membranes. Proteins were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce). The following primary antibodies were from Cell Signaling (all at a dilution of 1:1,000): anti-mTOR (2972); antibody to mTOR phosphorylated at Ser2448 (2971); anti-Akt (9272); antibody to Akt phosphorylated at Thr308 (9275); anti-4E-BP1 (9452); antibody to 4E-BP1 phosphorylated at Thr37 and Thr46 (9459), Ser65 (9451) or Thr70 (9455); anti-p70S6K (9202); and antibody to p70S6K phosphorylated at Thr389 (9205). Anti-β-actin was from Sigma. Anti-IRF7 (sc-9083) was from Santa Cruz. Horseradish peroxidase–conjugated secondary antibody (1:1,000) was from Cell Signaling.
Immunoblotting.
RAW cells (4 × 106) were transfected for 36 h with hemagglutinin-tagged MyD88 and YFP-tagged TLR9 or GFP-tagged TLR9 plasmids. Cells were pretreated with rapamycin (100 nM) for 3 h before being incubated with CpG-A–DOTAP for an additional 90 min. After being washed with cold PBS, cells were lysed and equal amounts of proteins from each sample were precleared for 1 h at 4 °C with immobilized protein A/G (Pierce). Supernatants were collected and were incubated overnight at 4 °C with anti-hemagglutinin (16B12; Covance) or anti-GFP (JL-8; Clontech) and immobilized protein A/G. Immune complex beads were washed thoroughly with lysis buffer. Sample buffer (30 μl) was added to the pellets and the proteins were released from the beads by boiling for 5 min. Coimmunoprecipitated proteins were detected by immunoblot analysis with anti-YFP or anti-hemagglutinin.
Additional methods.
Information on mouse injections, encapsulation of rapamycin in PLGA microparticles, apoptosis assays for pDCs, cytokine detection, flow cytometry, microarray analysis, imaging and luciferase reporter assays is available in the Supplementary Methods online.
Statistics.
The statistical significance of differences in the mean ± s.d. of cytokines released by cells of various groups was calculated with the Student's t-test (one tailed). P values less than 0.05 were considered statistically significant.
Note: Supplementary information is available on the Nature Immunology website.
References
Liu, Y.J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23, 275–306 (2005).
Colonna, M., Trinchieri, G. & Liu, Y.J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5, 1219–1226 (2004).
Asselin-Paturel, C. et al. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 2, 1144–1150 (2001).
Kawai, T. & Akira, S. Innate immune recognition of viral infection. Nat. Immunol. 7, 131–137 (2006).
Ito, T., Wang, Y.H. & Liu, Y.J. Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9. Springer Semin. Immunopathol. 26, 221–229 (2005).
Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 (2005).
Honda, K. et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 (2005).
Hazeki, K., Nigorikawa, K. & Hazeki, O. Role of phosphoinositide 3-kinase in innate immunity. Biol. Pharm. Bull. 30, 1617–1623 (2007).
Dann, S.G., Selvaraj, A. & Thomas, G. mTOR Complex1–S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol. Med. 13, 252–259 (2007).
Mamane, Y., Petroulakis, E., LeBacquer, O. & Sonenberg, N. mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 (2006).
Sarbassov, D.D., Ali, S.M. & Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17, 596–603 (2005).
Young, D.A. & Nickerson-Nutter, C.L. mTOR–beyond transplantation. Curr. Opin. Pharmacol. 5, 418–423 (2005).
Sehgal, S.N. Rapamune (RAPA, rapamycin, sirolimus): mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin. Biochem. 31, 335–340 (1998).
Krieg, A.M. Now I know my CpGs. Trends Microbiol. 9, 249–252 (2001).
Sigal, N.H., Dumont, F.J. & Cyclosporin, A. FK-506, and rapamycin: pharmacologic probes of lymphocyte signal transduction. Annu. Rev. Immunol. 10, 519–560 (1992).
Schreiber, S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251, 283–287 (1991).
Turnquist, H.R. et al. Rapamycin-conditioned dendritic cells are poor stimulators of allogeneic CD4+ T cells, but enrich for antigen-specific Foxp3+ T regulatory cells and promote organ transplant tolerance. J. Immunol. 178, 7018–7031 (2007).
Janus, A., Robak, T. & Smolewski, P. The mammalian target of the rapamycin (mTOR) kinase pathway: its role in tumourigenesis and targeted antitumour therapy. Cell. Mol. Biol. Lett. 10, 479–498 (2005).
Kozma, S.C. & Thomas, G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24, 65–71 (2002).
Richter, J.D. & Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 (2005).
Hay, N. & Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 (2004).
Nave, B.T., Ouwens, M., Withers, D.J., Alessi, D.R. & Shepherd, P.R. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem. J. 344, 427–431 (1999).
Colina, R. et al. Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008).
Latz, E. et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198 (2004).
Kawai, T. et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5, 1061–1068 (2004).
Sato, M. et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13, 539–548 (2000).
Marie, I., Smith, E., Prakash, A. & Levy, D.E. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20, 8803–8814 (2000).
Monath, T.P. Yellow fever vaccine. Expert Rev. Vaccines 4, 553–574 (2005).
Querec, T. et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 203, 413–424 (2006).
Singh, M., Chakrapani, A. & O'Hagan, D. Nanoparticles and microparticles as vaccine-delivery systems. Expert Rev. Vaccines 6, 797–808 (2007).
Shinohara, M.L. et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat. Immunol. 7, 498–506 (2006).
Mamane, Y. et al. eIF4E–from translation to transformation. Oncogene 23, 3172–3179 (2004).
Guiducci, C. et al. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J. Exp. Med. 205, 315–322 (2008).
Yamamoto, M. et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Kumar, H., Koyama, S., Ishii, K.J., Kawai, T. & Akira, S. Cutting edge: cooperation of IPS-1- and TRIF-dependent pathways in poly IC-enhanced antibody production and cytotoxic T cell responses. J. Immunol. 180, 683–687 (2008).
Waibler, Z. et al. Modified vaccinia virus Ankara induces Toll-like receptor-independent type I interferon responses. J. Virol. 81, 12102–12110 (2007).
Gallucci, S. & Matzinger, P. Danger signals: SOS to the immune system. Curr. Opin. Immunol. 13, 114–119 (2001).
Wilson, N.S. et al. Most lymphoid organ dendritic cell types are phenotypically and functionally immature. Blood 102, 2187–2194 (2003).
Proud, C.G. The multifaceted role of mTOR in cellular stress responses. DNA Repair (Amst.) 3, 927–934 (2004).
Banchereau, J. & Pascual, V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25, 383–392 (2006).
Acknowledgements
We thank S. Aguilar Mertens and L. Bronner for assistance with cell sorting; D. Kalman for assistance with deconvolution microscopy; K. Schaefer-Hales for assistance with confocal microscopy; T. Querec for YF-17D virus preparation; T. Denning for data discussion and technical advice; R. Nair for in vivo experiments; Y. Wang for immunofluorescence staining; J. Jiang for plasmid preparation; T. Taniguchi (University of Tokyo) for cyan fluorescent protein–tagged MyD88 and YFP-tagged IRF7 plasmids; D. Segal (National Cancer Institute) for GFP-tagged TLR9; D. Golenbock (University of Massachusetts) for the YFP-tagged TLR9 plasmid; B. Beutler (The Scripps Research Institute) for the hemagglutinin-tagged MyD88 plasmid; J. Hiscott (McGill University) for constitutively active IRF7; R. Ahmed (Emory University) for YF-17D subpassaged from YF-VAX in SW-480 cells; R.R. Amara (Emory University) for modified vaccinia virus Ankara; T. Van Dyke (Boston University) for highly purified E. coli LPS (strain 25922); S. Zughaier (Emory University) for the RAW264.7 cell line; and S.C. Kozma (Friedrich Miescher Institute for Biomedical Research) and N. Sonenberg (McGill University) for Rps6k1−/− Rps6k2−/− double-knockout mice. Supported by the National Institutes of Health (AI0564499, AI048638, AI05726601, DK057665, AI057157 and AI-50019 to B.P.)
Author information
Authors and Affiliations
Contributions
W.C. and B.P. designed the experiments; W.C. did the experiments; S.M. did the siRNA transfection; H.T. did the in vivo experiments; S.P.K. and N.M. prepared the microparticles; A.P. analyzed the microarray data; and W.C. and B.P. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Methods (PDF 451 kb)
Rights and permissions
About this article
Cite this article
Cao, W., Manicassamy, S., Tang, H. et al. Toll-like receptor–mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 9, 1157–1164 (2008). https://doi.org/10.1038/ni.1645
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1645
This article is cited by
-
Chemoproteomic development of SLC15A4 inhibitors with anti-inflammatory activity
Nature Chemical Biology (2024)
-
First-line steroid-free systemic treatment of acute and chronic graft-versus-host disease after novel prophylaxis regimens
Bone Marrow Transplantation (2023)
-
Vaccine adjuvants: mechanisms and platforms
Signal Transduction and Targeted Therapy (2023)
-
Regulation of the nucleic acid-sensing Toll-like receptors
Nature Reviews Immunology (2022)
-
SARS-CoV-2 pathogenesis
Nature Reviews Microbiology (2022)