Abstract
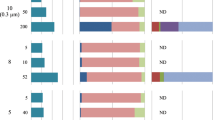
Deep-sea sediments harbour a vast biosphere. Archaea—one of the three domains of life1—are prevalent in marine environments2,3,4,5, and comprise a significant fraction of the biomass in marine sediments6. Archaeal membranes are well characterized, and are comprised of a glycerol backbone and a nonpolar isoprenoid chain. However, the ecology of sedimentary archaea remains elusive, because it is difficult to grow them in the laboratory. Here, we trace the fate of 13C-labelled glucose added to marine sediments in Sagami Bay, Japan, to determine the in situ mechanisms of membrane synthesis. Following the addition of labelled glucose to sediment samples collected in the region, we placed the cores on the sea floor and sampled them after 9 and 405 days. We found that the 13C was incorporated into the glycerol backbone of archaeal membranes; 13C was apparent after 9 days of incubation, but most pronounced after 405 days. However, the isoprenoid chain of the membranes remained unlabelled. On the basis of the differential uptake of 13C, we suggest that the glycerol unit is synthesized de novo, whereas the isoprenoid unit is synthesized from relic archaeal membranes and detritus, because of the prevalence of these compounds in marine sediments. We therefore suggest that some benthic archaea build their membranes by recycling sedimentary organic compounds.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms—proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579 (1990).
DeLong, E. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA 89, 5685–5689 (1992).
Ouverney, C. C. & Fuhrman, J. A. Marine planktonic Archaea take up amino acids. Appl. Environ. Microbiol. 66, 4829–4833 (2000).
Wuchter, C., Schouten, S., Boschker, H. T. S. & Sinninghe Damste, J. S. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219, 203–207 (2003).
Karner, M. B., DeLong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001).
Lipp, J. S., Morono, Y., Inagaki, F. & Hinrichs, K-U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994 (2008).
Parkes, R. J., Cragg, B. A. & Wellsbury, P. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol. J. 8, 11–28 (2000).
Rothschild, L. J. & Mancinelli, R. L. Life in extreme environments. Nature 409, 1092–1101 (2001).
D’Hondt, S. et al. Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221 (2004).
Schippers, A. et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 861–864 (2005).
Jorgensen, B. & Boetius, A. Feast and famine-microbial life in the deep-sea bed. Nature Rev. Microbiol. 5, 770–781 (2007).
Valentine, D. L. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nature Rev. Microbiol. 5, 316–323 (2007).
Inagaki, F. et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl Acad. Sci. USA 103, 2815–2820 (2006).
Roussel, E. G. et al. Extending the sub-sea-floor biosphere. Science 320, 1046–1046 (2008).
Biddle, J. F. et al. Heterotrophic archaea dominate sedimentary subsurface ecosystems of Peru. Proc. Natl Acad. Sci. USA 103, 3846–3851 (2006).
Teske, A. & Sorensen, K. B. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2, 3–18 (2008).
Nomaki, H., Heinz, P., Nakatsuka, T., Shimanaga, M. & Kitazato, H. Species-specific ingestion of organic carbon by deep-sea benthic foraminifera and meiobenthos: In situ tracer experiments. Limnol. Oceanogr. 50, 134–146 (2005).
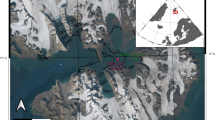
Kitazato, H. ‘The project Sagami’—dynamic sedimentary processes of both organic and inorganic materials at continental margins with active tectonic forcing. Prog. Oceanogr. 57, 1–2 (2003).
Shah, S. R., Mollenhauer, G., Ohkouchi, N., Eglinton, T. I. & Pearson, A. Origins of archaeal tetraether lipids in sediments: Insights from radiocarbon analysis. Geochim. Cosmochim. Acta 72, 4577–4594 (2008).
Koga, Y. & Morii, H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 71, 97–120 (2007).
Verhees, C. H. et al. The unique features of glycolytic pathways in archaea. Biochem. J. 375, 231–246 (2003).
Grochowski, L., Xu, H. & White, R. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188, 3192–3198 (2006).
Lodish, H. et al. Molecular Cell Biology (W. H. Freeman & Co, 1995).
Price, P. B. & Sowers, T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl Acad. Sci. USA 101, 4631–4636 (2004).
White, G., Russell, N. & Tidswell, E. Bacterial scission of ether bonds. Microbiol. Rev. 60, 216–232 (1996).
Hopmans, E. C., Schouten, S., Pancost, R. D., van der Meer, M. T. J. & Sinninghe Damste, J. S. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 14, 585–589 (2000).
Schouten, S., Hoefs, M. J. L., Koopmans, M. P., Bosch, H. J. & Sinninghe Damste, J. S. Structural characterization, occurrence and fate of archaeal ether-bound acyclic and cyclic biphytanes and corresponding diols in sediments. Org. Geochem. 29, 1305–1319 (1998).
Elvert, M., Suess, E., Greinert, J. & Whiticar, M. J. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org. Geochem. 31, 1175–1187 (2000).
Lane, D. J. et al. Rapid-determination of 16S ribosomal-RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA 82, 6955–6959 (1985).
Ludwig, W. et al. ARB: A software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004).
Acknowledgements
The authors thank T. Terada (Marine Works Japan), the shipboard scientists for their experimental and logistic support during cruises NT06-04,-05,-22 and NT08-02 by R/V Natsushima and for providing bathymetric data. This research was supported in part by a grant from the Japan Society for the Promotion of Science, Grant-in-Aid for Creative Scientific Research (19GS0211 & 22684030) and the internship program between Japan and Germany (FY2009).
Author information
Authors and Affiliations
Contributions
Y.T. carried out the lipids analysis and wrote the paper; Y.C. and N.O.O. supported the carbon isotope standard reagents and isotope ratio mass spectrometry analysis; H.N. and H.K. supported the 13C-substrate set-up, in situ deployment of the chamber, and core processing during cruises NT06-04, -05, -22 and NT08-02; Y.M. and F.I. supported phylogenic molecular analysis of 16S rRNA and qPCR; K-U.H. contributed to technical aspects and was involved in the study design; Y.T. and N.O. contributed to this study and all authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2236 kb)
Rights and permissions
About this article
Cite this article
Takano, Y., Chikaraishi, Y., Ogawa, N. et al. Sedimentary membrane lipids recycled by deep-sea benthic archaea. Nature Geosci 3, 858–861 (2010). https://doi.org/10.1038/ngeo983
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ngeo983
This article is cited by
-
Catabolic protein degradation in marine sediments confined to distinct archaea
The ISME Journal (2022)
-
Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria
The ISME Journal (2020)
-
Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years
Nature Communications (2020)
-
Environmental lipidomics: understanding the response of organisms and ecosystems to a changing world
Metabolomics (2020)
-
Insight into anaerobic methanotrophy from 13C/12C- amino acids and 14C/12C-ANME cells in seafloor microbial ecology
Scientific Reports (2018)