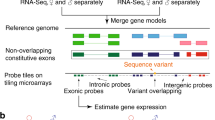
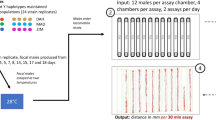
Abstract
Identifying the genes involved in polygenic traits has been difficult. In the 1950s and 1960s, laboratory selection experiments for extreme geotaxic behavior in fruit flies established for the first time that a complex behavioral trait has a genetic basis. But the specific genes responsible for the behavior have never been identified using this classical model. To identify the individual genes involved in geotaxic response, we used cDNA microarrays to identify candidate genes and assessed fly lines mutant in these genes for behavioral confirmation. We have thus determined the identities of several genes that contribute to the complex, polygenic behavior of geotaxis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Erlenmeyer-Kimling, L. & Hirsch, J. Measurement of the relations between chromosomes and behavior. Science 134, 1068–1069 (1961).
Hirsch, J. & Erlenmeyer-Kimling, L. Sign of taxis as a property of the genotype. Science 134, 835–836 (1961).
Erlenmeyer-Kimling, L., Hirsch, J. & Weiss, J.M. Studies in experimental behavior genetics: III. Selection and hybridization analyses of individual differences in the sign of geotaxis. J. Comp. Physiol. Psychol. 55, 722–731 (1962).
Hirsch, J. & Erlenmeyer-Kimling, L. Studies in experimental behavior genetics: IV. Chromosome analyses for geotaxis. J. Comp. Physiol. Psychol. 55, 732–739 (1962).
Ricker, J.P. & Hirsch, J. Evolution of an instinct under long-term divergent selection for geotaxis in domesticated populations of Drosophila melanogaster. J. Comp. Psych. 99, 380–390 (1985).
Dobzhansky, T. & Spassky, B. An experiment on migration and simultaneous selection for several traits in Drosophila pseudoobscura. Genetics 55, 723–734 (1966).
Dobzhansky, T., Spassky, B. & Sved, J. Effects of selection and migration on geotactic and phototactic behavior of Drosophila. II. Proc. R. Soc. Lond. B. 173, 191–207 (1968).
Dobzhansky, T., Levene, H. & Spassky, B. Effects of selection and migration on geotactic and phototactic behavior of Drosophila. III. Proc. R. Soc. Lond. B. 180, 21–41 (1972).
Stoltenberg, S.F. & Hirsch, J. A gene correlate of geotaxis near Adh (2–50.1) in Drosophila melanogaster. J. Comp. Psych. 110, 252–259 (1996).
Tully, T. Discovery of genes involved in learning and memory: an experimental synthesis of Hirschian and Benzerian perspectives. Proc. Natl Acad. Sci. USA 93, 13460–13467 (1996).
White, K.P. Functional genomics and the study of development, variation, and evolution. Nature Rev. Genet. 2, 528–537 (2001).
Ricker, J.P. & Hirsch, J. Reversal of genetic homeostasis in laboratory populations of Drosophila melanogaster under long-term selection for geotaxis and estimates of gene correlates: evolution of behavior-genetic systems. J. Comp. Psych. 102, 203–214 (1988).
White, K.P., Rifkin, S.A., Hurban, P. & Hogness, D.S. Microarray analysis of Drosophila development during metamorphosis. Science 286, 2179–2184 (1999).
Stanewsky, R. et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692 (1998).
Torok, I. et al. The overgrown hematopoietic organs-31 tumor suppressor gene of Drosophila encodes an Importin-like protein accumulating in the nucleus at the onset of mitosis. J. Cell Biol. 129, 1473–1489 (1995).
Renn, S.C., Park, J.H., Rosbash, M., Hall, J.C. & Taghert, P.H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999).
Doe, C.Q., Chu-LaGraff, Q., Wright, D.M. & Scott, M.P., The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464 (1991).
Roch, F. et al. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: clonal analysis and morphology of imaginal discs. Mol. Gen. Genet. 257, 103–112 (1998).
Zinsmaier, K.E., Eberle, K.K., Buchner, E., Walter, N. & Benzer, S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science 263, 977–980 (1994).
Osborne, K.A. et al. Natural behavioral polymorphism due to a cGMP-dependnet protein kinase of Drosophila. Science 277, 834–836 (1997).
Lin, Y.J., Seroude, L. & Benzer, S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282, 943–946 (1998).
Song, W. et al. Methuselah, a putative G protein-coupled receptor, regulates excitatory neurotransmitter exocytosis at the larval neuromuscular junction of Drosophila. A. Dros. Res. Conf. 42, 51 (2001).
Choi, K.-W. & Benzer, S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell 78, 125–136 (1994).
Konopka, R.J. & Benzer, S. Clock mutants of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 68, 2112–2116 (1971).
Kussel, P. & Frasch, M. Pendulin, a Drosophila protein with cell cycle-dependent nuclear localization, is required for normal cell proliferation. J. Cell Biol. 129, 1491–1507 (1995).
Emery, P., So, W.V., Kaneko, M., Hall, J.C. & Rosbash, M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 (1998).
Melzig, J., Burg, M., Gruhn, M., Pak, W.L. & Buchner, E. Selective histamine uptake rescues photo- and mechanoreceptor function of histidine decarboxylase-deficient Drosophila mutant. J. Neurosci. 18, 7160–7166 (1998).
Claridge-Chang, A. et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671 (2001).
McDonald, M.J. & Rosbash, M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578 (2001).
Greenspan, R.J. A kinder, gentler genetic analysis of behavior: dissection gives way to modulation. Curr. Opin. Neurobiol. 7, 805–811 (1997).
McMillan, P.A. & McGuire, T.R. The homeotic gene spineless-aristapedia affects geotaxis in Drosophila melanogaster. Behav. Genet. 22, 557–573 (1992).
Shaw, P.J., Cirelli, C., Greenspan, R.J. & Tononi, G. Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 (2000).
Acknowledgements
We thank R.W. Cross for help with statistical analyses, S. Bekele and D. Robinson for technical assistance and H. Dierick, G. Robinson and B. van Swinderen for reviewing the manuscript. The work by K.P.W. was supported by the National Institutes of Health, and all work at the Neurosciences Institute was supported by the Neurosciences Research Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Toma, D., White, K., Hirsch, J. et al. Identification of genes involved in Drosophila melanogaster geotaxis, a complex behavioral trait. Nat Genet 31, 349–353 (2002). https://doi.org/10.1038/ng893
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng893
This article is cited by
-
Essential elements of radical pair magnetosensitivity in Drosophila
Nature (2023)
-
No evidence for magnetic field effects on the behaviour of Drosophila
Nature (2023)
-
Analysis of negative phototaxis in the pill bug (Armadillidium vulgare) using omnidirectional servosphere
Artificial Life and Robotics (2023)
-
Phototransduction and circadian entrainment are the key pathways in the signaling mechanism for the baculovirus induced tree-top disease in the lepidopteran larvae
Scientific Reports (2018)
-
Inhalation toxicity of indoor air pollutants in Drosophila melanogaster using integrated transcriptomics and computational behavior analyses
Scientific Reports (2017)