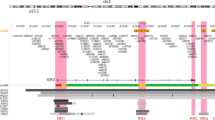
Abstract
We conducted a genome-wide association study (GWAS) of breast cancer by genotyping 528,173 SNPs in 1,145 postmenopausal women of European ancestry with invasive breast cancer and 1,142 controls. We identified four SNPs in intron 2 of FGFR2 (which encodes a receptor tyrosine kinase and is amplified or overexpressed in some breast cancers) that were highly associated with breast cancer and confirmed this association in 1,776 affected individuals and 2,072 controls from three additional studies. Across the four studies, the association with all four SNPs was highly statistically significant (Ptrend for the most strongly associated SNP (rs1219648) = 1.1 × 10−10; population attributable risk = 16%). Four SNPs at other loci most strongly associated with breast cancer in the initial GWAS were not associated in the replication studies. Our summary results from the GWAS are available online in a form that should speed the identification of additional risk loci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pharoah, P.D. et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 31, 33–36 (2002).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
Risch, H.A. et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J. Natl. Cancer Inst. 98, 1694–1706 (2006).
Hirschhorn, J.N. & Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Easton, D.F. et al. A genome-wide association study identifies multiple breast cancer susceptibility loci. Nature, advance online publication 27 May 2007 (doi:10.1038/nature05887).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 (2007).
Gudmundsson, J. et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 39, 631–637 (2007).
Tworoger, S.S., Eliassen, A.H., Sluss, P. & Hankinson, S.E. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J. Clin. Oncol. 25, 1482–1488 (2007).
Grose, R. & Dickson, C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 16, 179–186 (2005).
Minichiello, M.J. & Durbin, R. Mapping trait loci by use of inferred ancestral recombination graphs. Am. J. Hum. Genet. 79, 910–922 (2006).
Eliassen, A.H., Tworoger, S.S., Mantzoros, C.S., Pollak, M.N. & Hankinson, S.E. Circulating insulin and c-peptide levels and risk of breast cancer among predominately premenopausal women. Cancer Epidemiol. Biomarkers Prev. 16, 161–164 (2007).
Hayes, R.B. et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control. Clin. Trials 21, 349S–355S (2000).
Stevens, V.L., Rodriguez, C., Pavluck, A.L., Thun, M.J. & Calle, E.E. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II Nutrition Cohort. Cancer Epidemiol. Biomarkers Prev. 15, 1226–1228 (2006).
Bruzzi, P., Green, S.B., Byar, D.P., Brinton, L.A. & Schairer, C. Estimating the population attributable risk for multiple risk factors using case-control data. Am. J. Epidemiol. 122, 904–914 (1985).
Moffa, A.B. & Ethier, S.P. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J. Cell. Physiol. 210, 720–731 (2007).
Chanock, S.J. et al. What constitutes replication of a genotype-phenotype association? Summary of an NCI-NHGRI working group. Nature (in the press).
Wang, H., Thomas, D.C., Pe'er, I. & Stram, D.O. Optimal two-stage genotyping designs for genome-wide association scans. Genet. Epidemiol. 30, 356–368 (2006).
Falush, D., Stephens, M. & Pritchard, J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Kraft, P., Cox, D.G., Paynter, R.A., Hunter, D. & De Vivo, I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet. Epidemiol. 28, 261–272 (2005).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Patterson, N., Price, A.L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Acknowledgements
We thank B. Egan, L. Egan, H. Judge Ellis, H. Ranu and P. Soule for assistance, and we thank the participants in the Nurses' Health Studies. We thank P. Prorok (Division of Cancer Prevention, National Cancer Institute); the Screening Center investigators and staff of PLCO; T. Riley, C. Williams and staff (Information Management Services, Inc.); B. O'Brien and staff (Westat, Inc.) and B. Kopp, T. Sheehy and staff (SAIC-Frederick). We acknowledge the study participants for their contributions in making this study possible. We thank C. Lichtman for data management and the participants on the CPS-II. We thank M. Minichiello for providing the Margarita program and for discussions. We acknowledge D. Easton and colleagues for sharing prepublication results. The Nurses' Health Studies are supported by US NIH grants CA65725, CA87969, CA49449, CA67262, CA50385 and 5UO1CA098233. The ACS study is supported by UO1 CA098710. The PLCO study is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Distribution of the admixture vector of each NHS participant as determined by STRUCTURE. (PDF 41 kb)
Supplementary Fig. 2
Probability-probability plot of the uncorrected P values (blue dots) compared with the expected uniform distribution (green line), where magenta dots show the P values corrected for age in 5-year intervals, an indicator for recent hormone use, and three eigenvectors controlling for population stratification. (PDF 77 kb)
Supplementary Fig. 3
ARG analysis with 81 SNPs after 106 permutations. (PDF 55 kb)
Supplementary Table 1
Identification of protective and at-risk haplotypes for the FGFR2 susceptibility locus. (PDF 79 kb)
Supplementary Table 2
Evidence for association between the six most significant SNPs in the NHS genome-wide association scan, the three replication studies, and the pooled scan and replication data. (PDF 56 kb)
Rights and permissions
About this article
Cite this article
Hunter, D., Kraft, P., Jacobs, K. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39, 870–874 (2007). https://doi.org/10.1038/ng2075
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2075
This article is cited by
-
Genome-wide association testing in malaria studies in the presence of overdominance
Malaria Journal (2023)
-
Identifying Lymph Node Metastasis-Related Factors in Breast Cancer Using Differential Modular and Mutational Structural Analysis
Interdisciplinary Sciences: Computational Life Sciences (2023)
-
Association of ABCC4 G559T single nucleotide polymorphism with arsenic-induced precancerous hyperkeratosis
The Nucleus (2023)
-
Heterozygotic Brca1 mutation initiates mouse genome instability at embryonic stage
Oncogenesis (2022)
-
Induced mammary cancer in rat models: pathogenesis, genetics, and relevance to female breast cancer
Journal of Mammary Gland Biology and Neoplasia (2022)