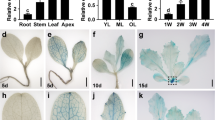
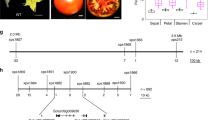
Abstract
Plant leaves show pronounced plasticity of size and form. In the classical, partially dominant mutation Lanceolate (La)1, the large compound leaves of tomato (Solanum lycopersicum) are converted into small simple ones. We show that LA encodes a transcription factor from the TCP family2,3 containing an miR319-binding site4. Five independent La isolates are gain-of-function alleles that result from point mutations within the miR319-binding site and confer partial resistance of the La transcripts to microRNA (miRNA)-directed inhibition. The reduced sensitivity to miRNA regulation leads to elevated LA expression in very young La leaf primordia and to precocious differentiation of leaf margins. In contrast, downregulation of several LA-like genes using ectopic expression of miR319 resulted in larger leaflets and continuous growth of leaf margins. Our results imply that regulation of LA by miR319 defines a flexible window of morphogenetic competence along the developing leaf margin that is required for leaf elaboration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mathan, D.S. & Jenkins, J.A. Chemically induced phenocopy of a tomato mutant. Science 131, 36–87 (1960).
Cubas, P., Lauter, N., Doebley, J. & Coen, E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222 (1999).
Nath, U., Crawford, B.C., Carpenter, R. & Coen, E. Genetic control of surface curvature. Science 299, 1404–1407 (2003).
Palatnik, J.F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 (2003).
Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y. & Lifschitz, E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84, 735–744 (1996).
Holtan, H.E. & Hake, S. Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165, 1541–1550 (2003).
Kessler, S., Kim, M., Pham, T., Weber, N. & Sinha, N. Mutations altering leaf morphology in tomato. Int. J. Plant Sci. 162, 475–492 (2001).
Menda, N., Semel, Y., Peled, D., Eshed, Y. & Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 38, 861–872 (2004).
Caruso, J.L. Morphogenetic aspects of a leafless mutant in tomato. I. General patterns in development. Am. J. Bot. 55, 1169–1176 (1968).
Dengler, N.G. Comparison of leaf development in Normal (+/+), Entire (E/E), and Lanceolate (La/+) plants of tomato, Lycopersicon esculentum 'Ailsa Craig'. Bot. Gaz. 145, 66–77 (1984).
Mathan, D.S. & Jenkins, J.A. A morphogenetic study of Lanceolate, a leaf shape mutant in the tomato. Am. J. Bot. 49, 504–514 (1962).
Stettler, R.F. Dosage effects of the Lanceolate gene in tomato. Am. J. Bot. 51, 253–264 (1964).
Hagemann, W. & Gleissberg, S. Organogenetic capacity of leaves: the significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199, 121–152 (1996).
Dengler, N.G. & Tsukaya, H. Leaf morphogenesis in dicotyledons: current issues. Int. J. Plant Sci. 162, 459–464 (2001).
Boynton, J.E. & Rick, C.M. Linkage tests with mutants of Stubbe's groups I, II, III and IV. Rep. Tomato Genet. Coop. Tomato Genet. Coop. 15, 24–27 (1965).
Li, C., Potuschak, T., Colon-Carmona, A., Gutierrez, R.A. & Doerner, P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102, 12978–12983 (2005).
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P. & Burge, C.B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
Schwab, R. et al. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527 (2005).
Lifschitz, E. et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103, 6398–6403 (2006).
Lifschytz, E. & Falk, R. A genetic analysis of the Killer-prune (K-pn) locus of Drosophila melanogaster. Genetics 62, 353–358 (1969).
Stadler, L.J. Some genetic effects of X rays in plants. J. Hered. 21, 3–19 (1930).
Chen, J.-J., Janssen, B.-J., Williams, A. & Sinha, N. A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9, 1289–1304 (1997).
Parnis, A. et al. The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9, 2143–2158 (1997).
Alvarez, J.P. et al. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18, 1134–1151 (2006).
Baker, C.C., Sieber, P., Wellmer, F. & Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303–315 (2005).
McConnell, J.R. et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 (2001).
Tang, G., Reinhart, B.J., Bartel, D.P. & Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63 (2003).
Bharathan, G. et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science 296, 1858–1860 (2002).
Hay, A. & Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38, 942–947 (2006).
Valoczi, A., Varallyay, E., Kauppinen, S., Burgyan, J. & Havelda, Z. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47, 140–151 (2006).
Acknowledgements
We wish to thank E. Lifschitz5 (Technion), the C.M. Rick Tomato Genetics Resource Center and Clemson University Genomics Institute for materials, Navot Ori for unearthing la-6, and R. Fluhr, E. Hornstein, Z. Lippman, D. Weigel, S. Hake, J. Bowman and members of our laboratories for discussions and criticism. The work was supported by grants from the US-Israel Binational Agricultural Research and Development Fund, the European Union (MechPlant project) and the Israel Science Foundation (ISF) to N.O.; from ISF, MINERVA, The US-Israel Binational Science Foundation and Lubin Center for Plant Biotechnology to Y.E.; from the German-Israeli Foundation for Scentific Research and Development to Y.E. and N.O.; and from the German-Israeli Project Cooperation to Y.E. and D.Z.
Author information
Authors and Affiliations
Contributions
All authors contributed to the experiments and their interpretation. N.O., D.Z. and Y.E. directed the experiments, and N.O. and Y.E. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
The Lanceolate syndrome in leaves and flowers. (PDF 619 kb)
Supplementary Fig. 2
Genetic and molecular characterization of LA. (PDF 473 kb)
Supplementary Fig. 3
The LA-like protein family. (PDF 999 kb)
Supplementary Fig. 4
Quantification of LA and Tkn2/LeT6 expression in genotypes representing a gradient of LA activity. (PDF 358 kb)
Supplementary Fig. 5
Heterochronic differences in wild-type and la-6 leaf morphology. (PDF 564 kb)
Supplementary Fig. 6
Genetic interactions of La-2 with other leaf shape mutants. (PDF 291 kb)
Supplementary Table 1
Primers used in this study (PDF 19 kb)
Rights and permissions
About this article
Cite this article
Ori, N., Cohen, A., Etzioni, A. et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787–791 (2007). https://doi.org/10.1038/ng2036
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2036
This article is cited by
-
Cucumber mosaic virus-induced gene and microRNA silencing in water dropwort (Oenanthe javanica (Blume) DC)
Plant Methods (2024)
-
Whole transcriptome analysis and construction of a ceRNA regulatory network related to leaf and petiole development in Chinese cabbage (Brassica campestris L. ssp. pekinensis)
BMC Genomics (2023)
-
Control of compound leaf patterning by MULTI-PINNATE LEAF1 (MPL1) in chickpea
Nature Communications (2023)
-
Dissected Leaf 1 encodes an MYB transcription factor that controls leaf morphology in potato
Theoretical and Applied Genetics (2023)
-
Implications of small RNAs in plant development, abiotic stress response and crop improvement in changing climate
The Nucleus (2023)