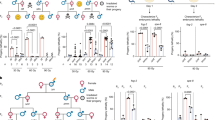
Abstract
There is increasing evidence that epigenetic information can be inherited across generations in mammals, despite extensive reprogramming both in the gametes and in the early developing embryo. One corollary to this is that disrupting the establishment of epigenetic state in the gametes of a parent, as a result of heterozygosity for mutations in genes involved in reprogramming, could affect the phenotype of offspring that do not inherit the mutant allele. Here we show that such effects do occur following paternal inheritance in the mouse. We detected changes to transcription and chromosome ploidy in adult animals. Paternal effects of this type have not been reported previously in mammals and suggest that the untransmitted genotype of male parents can influence the phenotype of their offspring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Chong, S. & Whitelaw, E. Epigenetic germline inheritance. Curr. Opin. Genet. Dev. 14, 692–696 (2004).
Rakyan, V. & Whitelaw, E. Transgenerational epigenetic inheritance. Curr. Biol. 13, R6 (2003).
Fitch, K.R., Yasuda, G.K., Owens, K.N. & Wakimoto, B.T. Paternal effects in Drosophila: implications for mechanisms of early development. Curr. Top. Dev. Biol. 38, 1–34 (1998).
Payer, B. et al. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13, 2110–2117 (2003).
Blewitt, M.E. et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102, 7629–7634 (2005).
Locke, J., Kotarski, M.A. & Tartof, K.D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120, 181–198 (1988).
Spofford, J.B. in The Genetics and Biology of Drosophila Vol. 1c (ed. Ashburner, M.) 955–1018 (Academic, London, 1976).
Fiering, S., Whitelaw, E. & Martin, D.I. To be or not to be active: the stochastic nature of enhancer action. Bioessays 22, 381–387 (2000).
Rakyan, V.K., Blewitt, M.E., Druker, R., Preis, J.I. & Whitelaw, E. Metastable epialleles in mammals. Trends Genet. 18, 348–351 (2002).
Reuter, G. & Spierer, P. Position effect variegation and chromatin proteins. Bioessays 14, 605–612 (1992).
Dodge, J.E. et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 280, 17986–17991 (2005).
Gaudet, F. et al. Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003).
Peters, A.H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001).
Stopka, T. & Skoultchi, A.I. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA 100, 14097–14102 (2003).
Duhl, D.M., Vrieling, H., Miller, K.A., Wolff, G.L. & Barsh, G.S. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8, 59–65 (1994).
Morgan, H.D., Sutherland, H.G., Martin, D.I. & Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314–318 (1999).
Blewitt, M.E., Vickaryous, N.K., Paldi, A., Koseki, H. & Whitelaw, E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2, e49 (2006).
Gaudet, F. et al. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 24, 1640–1648 (2004).
Rakyan, V.K. et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 100, 2538–2543 (2003).
Wolff, G.L. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics 88, 529–539 (1978).
Tsukiyama, T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 3, 422–429 (2002).
Lazzaro, M.A. & Picketts, D.J. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 77, 1145–1156 (2001).
Rassoulzadegan, M. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006).
Aravin, A. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006).
Girard, A., Sachidanandam, R., Hannon, G.J. & Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006).
Grivna, S.T., Beyret, E., Wang, Z. & Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006).
Watanabe, T. et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743 (2006).
Torres-Padilla, M.E. & Zernicka-Goetz, M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J. Cell Biol. 174, 329–338 (2006).
Gruenbaum, Y., Cedar, H. & Razin, A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 295, 620–622 (1982).
Li, E., Bestor, T.H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
La Salle, S. et al. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 268, 403–415 (2004).
Bourc'his, D. & Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004).
Webster, K.E. et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. USA 102, 4068–4073 (2005).
Bourc'his, D., Xu, G.L., Lin, C.S., Bollman, B. & Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Takagi, N. & Abe, K. Detrimental effects of two active X chromosomes on early mouse development. Development 109, 189–201 (1990).
Cattanach, B.M. Genetic disorders of sex determination in mice and other mammals. in Birth Defects. Proceedings of the Fourth International Conference (eds. Motulsky, A. & Lenz, W.) 129–141 (Excerpta Medica, Amsterdam, 1974).
Christians, E., Davis, A.A., Thomas, S.D. & Benjamin, I.J. Maternal effect of Hsf1 on reproductive success. Nature 407, 693–694 (2000).
Dean, J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J. Reprod. Immunol. 53, 171–180 (2002).
Gurtu, V.E. et al. Maternal effect for DNA mismatch repair in the mouse. Genetics 160, 271–277 (2002).
Howell, C.Y. et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838 (2001).
Tong, Z.B. et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 26, 267–268 (2000).
Wu, X. et al. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 33, 187–191 (2003).
Szabad, J., Mathe, E. & Puro, J. Horka, a dominant mutation of Drosophila, induces nondisjunction and, through paternal effect, chromosome loss and genetic mosaics. Genetics 139, 1585–1599 (1995).
Sassone-Corsi, P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178 (2002).
van der Heijden, G.W. et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev. Biol. 298, 458–469 (2006).
Lane, N. et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003).
Cheutin, T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003).
McClive, P.J. & Sinclair, A.H. Rapid DNA extraction and PCR-sexing of mouse embryos. Mol. Reprod. Dev. 60, 225–226 (2001).
Li, J.Y., Lees-Murdock, D.J., Xu, G.L. & Walsh, C.P. Timing of establishment of paternal methylation imprints in the mouse. Genomics 84, 952–960 (2004).
Arnaud, P. et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12, 1005–1019 (2003).
Acknowledgements
This study was supported by National Health and Medical Research Council of Australia grants to E.W., D.D.K., H.S., J.M. and M.O'B. and an Australian Research Council grant to S.C. and E.W. M.B., A.A. and N.Z. were supported by Australian Postgraduate Awards. N.V. was supported by an International Postgraduate Award (University of Sydney). N.Y. and E.W. were supported by fellowships from the Queensland Institute of Medical Research.
Author information
Authors and Affiliations
Contributions
S.C., N.V., A.A., N.Z., N.Y., S.H. and M.B. performed experiments and provided intellectual input. T.S., A.S. and H.S.S. provided backcrossed mice. D.D.K., M.O'B., J.M., H.S.S. and E.W. provided intellectual input. S.C., N.V., A.A., N.Y. and E.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Homology model of mut-snf2h. (PDF 44 kb)
Supplementary Fig. 2
RNA blot analysis of IAP RNA in Smarca5Momme D4/+ testes. (PDF 59 kb)
Supplementary Fig. 3
DNA methylation analysis of repeats in Dnmt1Momme D2 embryos. (PDF 73 kb)
Supplementary Fig. 4
Methylation at the Mtm1 CpG island in mouse 1 and mouse 2. (PDF 115 kb)
Supplementary Fig. 5
Methylation analysis of DNA from the sperm of Dnmt1Momme D2/+ and Dnmt3l+/− mice. (PDF 120 kb)
Supplementary Table 1
Genes in the Momme D4 linked interval. (PDF 14 kb)
Supplementary Table 2
Smarca5Momme D4 phenotypes. (PDF 29 kb)
Supplementary Table 3
Primers. (PDF 14 kb)
Rights and permissions
About this article
Cite this article
Chong, S., Vickaryous, N., Ashe, A. et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet 39, 614–622 (2007). https://doi.org/10.1038/ng2031
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2031
This article is cited by
-
Generating specificity in genome regulation through transcription factor sensitivity to chromatin
Nature Reviews Genetics (2022)
-
BAF complex-mediated chromatin relaxation is required for establishment of X chromosome inactivation
Nature Communications (2022)
-
Genetic control of non-genetic inheritance in mammals: state-of-the-art and perspectives
Mammalian Genome (2020)
-
Exploring the extent and scope of epigenetic inheritance
Nature Reviews Endocrinology (2018)
-
Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications
Nature Reviews Genetics (2016)