Abstract
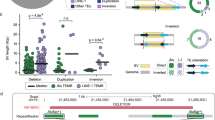
Several large-scale studies of human genetic variation have provided insights into processes such as recombination that have shaped human diversity. However, regions such as low-copy repeats (LCRs) have proven difficult to characterize, hindering efforts to understand the processes operating in these regions. We present a detailed study of genetic variation and underlying recombination processes in two copies of an LCR (NF1REPa and NF1REPc) on chromosome 17 involved in the generation of NF1 microdeletions and in a third copy (REP19) on chromosome 19 from which the others originated over 6.7 million years ago. We find evidence for shared hotspots of recombination among the LCRs. REP19 seems to contain hotspots in the same place as the nonallelic recombination hotspots in NF1REPa and NF1REPc. This apparent conservation of patterns of recombination hotspots in moderately diverged paralogous regions contrasts with recent evidence that these patterns are not conserved in less-diverged orthologous regions of chimpanzees.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Lopez-Correa, C. et al. Recombination hotspot in NF1 microdeletion patients. Hum. Mol. Genet. 10, 1387–1392 (2001).
Repping, S. et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 71, 906–922 (2002).
Reiter, L.T. et al. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am. J. Hum. Genet. 62, 1023–1033 (1998).
Lopez Correa, C., Brems, H., Lazaro, C., Marynen, P. & Legius, E. Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am. J. Hum. Genet. 66, 1969–1974 (2000).
Forbes, S.H., Dorschner, M.O., Le, R. & Stephens, K. Genomic context of paralogous recombination hotspots mediating recurrent NF1 region microdeletion. Genes Chromosom. Cancer 41, 12–25 (2004).
Petek, E. et al. Mitotic recombination mediated by the JJAZF1 (KIAA0160) gene causing somatic mosaicism and a new type of constitutional NF1 microdeletion in two children of a mosaic female with only few manifestations. J. Med. Genet. 40, 520–525 (2003).
Kehrer-Sawatzki, H. et al. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am. J. Hum. Genet. 75, 410–423 (2004).
Jeffreys, A.J. & May, C.A. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36, 151–156 (2004).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
Crawford, D.C. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36, 700–706 (2004).
Ptak, S.E. et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2, e155 (2004).
Fearnhead, P., Harding, R.M., Schneider, J.A., Myers, S. & Donnelly, P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics 167, 2067–2081 (2004).
Fearnhead, P. & Smith, N.G. A novel method with improved power to detect recombination hotspots from polymorphism data reveals multiple hotspots in human genes. Am. J. Hum. Genet. 77, 781–794 (2005).
Hudson, R.R. Two-locus sampling distributions and their application. Genetics 159, 1805–1817 (2001).
Jeffreys, A.J., Neumann, R., Panayi, M., Myers, S. & Donnelly, P. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 37, 601–606 (2005).
De Raedt, T. et al. Genomic organization and evolution of the NF1 microdeletion region. Genomics 84, 346–360 (2004).
Ptak, S.E. et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37, 429–434 (2005).
Winckler, W. et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308, 107–111 (2005).
Neumann, R. & Jeffreys, A.J. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum. Mol. Genet. 15, 1401–1411 (2006).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Stephens, M. & Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003).
Marchini, J. et al. A comparison of phasing algorithms for trios and unrelated individuals. Am. J. Hum. Genet. 78, 437–450 (2006).
Acknowledgements
The authors thank T. de Ravel for critically reading the manuscript. T.D. is supported by the Emmanuel Vanderschueren Fonds. M.S. is supported by US National Institutes of Health grant 1RO1HG/LM02585-01. E.L. is a part-time clinical researcher of the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO). This work is also supported by research grants from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (G.0096.02, E.L.; G.0507.04, P.M.); the Interuniversity Attraction Poles (IAP) granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium (2002-2006; P5/25) (P.M. and E.L.); by a Concerted Action Grant from the Catholic University of Leuven and by the Belgian Federation against Cancer (SCIE2003-33 to E.L.).
Author information
Authors and Affiliations
Contributions
This study was coordinated by T.D., P.M. and E.L.; the manuscript was written by T.D., M.S. and E.L.; breakpoint detection was performed by T.D., I.H., H.B., L.M., K.S., C.L., K.W., H.K., D.V. and L.K.; SNP detection and typing was performed by T.D., I.H., H.B. and D.T. and computational analysis was performed by T.D. and M.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Schematic overview of the NF1 microdeletion region. (PDF 492 kb)
Supplementary Fig. 2
LD pattern of the different LCRs. (PDF 416 kb)
Supplementary Fig. 3
Distribution and ratio of lambda values in the LCRs. (PDF 427 kb)
Supplementary Fig. 4
Breakpoint detection. (PDF 548 kb)
Supplementary Table 1
dbSNP numbers (ss) of SNPs used in shared SNP study. (PDF 72 kb)
Supplementary Table 2
SNPs used in the genotyping of 43 nuclear families (PDF 76 kb)
Supplementary Table 3
Primer list. (PDF 903 kb)
Supplementary Table 4
Haplotype and input data. (XLS 123 kb)
Rights and permissions
About this article
Cite this article
Raedt, T., Stephens, M., Heyns, I. et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet 38, 1419–1423 (2006). https://doi.org/10.1038/ng1920
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1920
This article is cited by
-
Assisted reproduction mediated resurrection of a feline model for Chediak-Higashi syndrome caused by a large duplication in LYST
Scientific Reports (2020)
-
Ultra-deep amplicon sequencing indicates absence of low-grade mosaicism with normal cells in patients with type-1 NF1 deletions
Human Genetics (2019)
-
Extreme clustering of type-1 NF1 deletion breakpoints co-locating with G-quadruplex forming sequences
Human Genetics (2018)
-
Pronounced maternal parent-of-origin bias for type-1 NF1 microdeletions
Human Genetics (2018)
-
Emerging genotype–phenotype relationships in patients with large NF1 deletions
Human Genetics (2017)