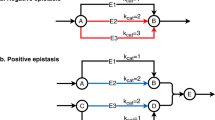
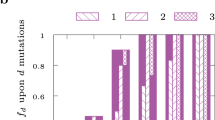
Abstract
Genetic robustness characterizes the constancy of the phenotype in face of heritable perturbations. Previous investigations have used comprehensive single and double gene knockouts to study gene essentiality and pairwise gene interactions in the yeast Saccharomyces cerevisiae. Here we conduct an in silico multiple knockout investigation of a flux balance analysis model of the yeast's metabolic network. Cataloging gene sets that provide mutual functional backup, we identify sets of up to eight interacting genes and characterize the 'k robustness' (the depth of backup interactions) of each gene. We find that 74% (360) of the metabolic genes participate in processes that are essential to growth in a standard laboratory environment, compared with only 13% previously found to be essential using single knockouts. The genes' k robustness is shown to be a solid indicator of their biological buffering capacity and is correlated with both the genes' environmental specificity and their evolutionary retention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
De Visser, J.A. et al. Perspective: evolution and detection of genetic robustness. Evolution Int. J. Org. Evolution 57, 1959–1972 (2003).
Kitano, H. Biological robustness. Nat. Rev. Genet. 5, 826–837 (2004).
Papp, B., Pál, C. & Hurst, L.D. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429, 661–664 (2004).
Stelling, J., Sauer, U., Szallasi, Z., Doyle, F.J. & Doyle, J. Robustness of cellular functions. Cell 118, 675–685 (2004).
Gu, Z. et al. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66 (2003).
Hartman, J.L., IV, Garvik, B. & Hartwell, L. Principles for the buffering of genetic variation. Science 291, 1001–1004 (2001).
Wagner, A. Robustness against mutations in genetic networks of yeast. Nat. Genet. 24, 355–361 (2000).
Nowak, M.A., Boerlijst, M.C., Cooke, J. & Maynard Smith, J. Evolution of genetic redundancy. Nature 388, 167–171 (1997).
Winzeler, E.A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Tong, A.H. et al. Global mapping of the yeast genetic interaction network. Science 303, 808–813 (2004).
Segrè, D., DeLuna, A., Church, G.M. & Kishony, R. Modular epistasis in yeast metabolism. Nat. Genet. 37, 77–83 (2004).
Thiele, I., Vo, T.D., Price, N.D. & Palsson, B.Ø. An expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single and double deletion mutants. J. Bacteriol. 187, 5818–5830 (2005).
Elena, S.F. & Lenski, R.E. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390, 395–398 (1997).
Klamt, S. & Gilles, E.D. Minimal cut sets in biochemical reaction networks. Bioinformatics 20, 226–234 (2004).
Schuster, S., Fell, D. & Dandekar, T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18, 326–332 (2000).
Förster, J., Famili, I., Palsson, B. Ø. & Nielsen, J. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. OMICS 7, 193–202 (2003).
Förster, J., Famili, I., Fu, P., Palsson, B. Ø. & Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13, 244–253 (2003).
Famili, I., Förster, J., Nielsen, J. & Palsson, B.Ø. Saccharomyces cerevisiae phenotypes can be predicted using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA 100, 13134–13139 (2003).
Kauffman, K.J., Prakash, P. & Edwards, J.S. Advances in flux balance analysis. Curr. Opin. Biotechnol. 14, 491–496 (2003).
Krylov, D.M., Wolf, Y.I., Rogozin, I.B. & Koonin, E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13, 2229–2235 (2003).
Wolf, Y.I., Carmel, L. & Koonin, E.V. Unifying measures of gene function and evolution. Proc. Biol. Sci. 273, 1507–1515 (2006).
Hirsh, A.E. & Fraser, H.B. Protein dispensability and rate of evolution. Nature 411, 1046–1049 (2001).
Papp, B., Pál, C. & Hurst, L.D. Genomic function (communication arising): rate of evolution and gene dispensability. Nature 421, 496–497 (2003).
Nelson, D.L. & Cox, M.M. Lehninger Principles of Biochemistry 3rd edn. (Worth Publishers, New York, 2000).
Schaaff-Gerstenschläger, I., Mannhaupt, G., Vetter, I., Zimmermann, F.K. & Feldmann, H. TKL2, a second transketolase gene of Saccharomyces cerevisiae – cloning, sequence and deletion analysis of the gene. Eur. J. Biochem. 217, 487–492 (1993).
Lord, P.W., Stevens, R., Brass, A. & Goble, C.A. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics 19, 1275–1283 (2003).
Blank, L.M., Kuepfer, L. & Sauer, U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 6, R49 (2005).
Meiklejohn, C.D. & Hartl, D.L. A single mode of canalization. Trends Ecol. Evol. 17, 468–473 (2002).
Benjamini, Y., Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Harbison, C.T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004).
Sharan, R. et al. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. USA 102, 1974–1979 (2005).
Hughes, T.R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Acknowledgements
We thank the Tauber fund for supporting D.D. Discussions with and comments of A. Hirsh, A. Kaufman, O. Meshi, Y. Pilpel, T. Pupko, R. Sharan, T. Shlomi and I. Venger are much appreciated. Figure 4 was drawn using Pajek from http://vlado.fmf.uni-lj.si/pub/networks/pajek/. M.K.'s work was supported by grants from the Israeli Science Foundation (ISF) and the Israeli Ministry of Health. E.R.'s research is supported by the Yishayahu Horowitz Center for Complexity Science, the Israeli Science Foundation (ISF), and the German-Israeli Foundation for scientific research and development (GIF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Proline and arginine metabolism (partial diagram). (PDF 32 kb)
Supplementary Fig. 2
Distribution of the number of essential gene sets per gene. (PDF 9 kb)
Supplementary Fig. 3
Functional distance values of coessential genes. (PDF 15 kb)
Supplementary Fig. 4
Growth rates of tested mutants missing two to four genes. (PDF 22 kb)
Supplementary Table 1
Model genes and the essential sets found. (PDF 60 kb)
Supplementary Table 2
Correlation of k robustness and several biological indicators. (PDF 25 kb)
Rights and permissions
About this article
Cite this article
Deutscher, D., Meilijson, I., Kupiec, M. et al. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet 38, 993–998 (2006). https://doi.org/10.1038/ng1856
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1856
This article is cited by
-
Local flux coordination and global gene expression regulation in metabolic modeling
Nature Communications (2023)
-
Integration of enzyme constraints in a genome-scale metabolic model of Aspergillus niger improves phenotype predictions
Microbial Cell Factories (2021)
-
Exploring gene knockout strategies to identify potential drug targets using genome-scale metabolic models
Scientific Reports (2021)
-
Genetic investigation of purine nucleotide imbalance in Saccharomyces cerevisiae
Current Genetics (2020)
-
Modeling the Interplay between Photosynthesis, CO2 Fixation, and the Quinone Pool in a Purple Non-Sulfur Bacterium
Scientific Reports (2019)