Abstract
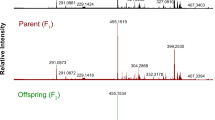
Variation for metabolite composition and content is often observed in plants. However, it is poorly understood to what extent this variation has a genetic basis. Here, we describe the genetic analysis of natural variation in the metabolite composition in Arabidopsis thaliana. Instead of focusing on specific metabolites, we have applied empirical untargeted metabolomics using liquid chromatography–time of flight mass spectrometry (LC-QTOF MS). This uncovered many qualitative and quantitative differences in metabolite accumulation between A. thaliana accessions. Only 13.4% of the mass peaks were detected in all 14 accessions analyzed. Quantitative trait locus (QTL) analysis of more than 2,000 mass peaks, detected in a recombinant inbred line (RIL) population derived from the two most divergent accessions, enabled the identification of QTLs for about 75% of the mass signals. More than one-third of the signals were not detected in either parent, indicating the large potential for modification of metabolic composition through classical breeding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wink, M. Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 75, 225–233 (1988).
Windsor, A.J. et al. Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae). Phytochemistry 66, 1321–1333 (2005).
Jansen, R.C. & Nap, J.P. Genetical genomics: the added value from segregation. Trends Genet. 17, 388–391 (2001).
Jansen, R.C. Interval mapping of multiple quantitative trait loci. Genetics 135, 205–211 (1993).
Bentsink, L. et al. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis . Plant Physiol. 124, 1595–1604 (2000).
Hobbs, D.H., Flintham, J.E. & Hills, M.J. Genetic control of storage oil synthesis in seeds of Arabidopsis . Plant Physiol. 136, 3341–3349 (2004).
Kliebenstein, D.J., Gershenzon, J. & Mitchell-Olds, T. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159, 359–370 (2001).
Loudet, O., Chaillou, S., Merigout, P., Talbotec, J. & Daniel-Vedele, F. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis . Plant Physiol. 131, 345–358 (2003).
Mita, S., Murano, N., Akaike, M. & Nakamura, K. Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 11, 841–851 (1997).
Tikunov, Y. et al. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139, 1125–1137 (2005).
Vorst, O. et al. A non-directed approach to the differential analysis of multiple LC-MS-derived metabolic profiles. Metabolomics 1, 169–180 (2005).
Dixon, R.A. Engineering of plant natural product pathways. Curr. Opin. Plant Biol. 8, 329–336 (2005).
Alonso-Blanco, C. et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14, 259–271 (1998).
Broman, K.W. Mapping quantitative trait loci in the case of a spike in the phenotype distribution. Genetics 163, 1169–1175 (2003).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
de Koning, D.J. & Haley, C.S. Genetical genomics in humans and model organisms. Trends Genet. 21, 377–381 (2005).
Mitchell-Olds, T. & Pedersen, D. The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis . Genetics 149, 739–747 (1998).
Reichelt, M. et al. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana . Phytochemistry 59, 663–671 (2002).
Kliebenstein, D.J. et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825 (2001).
Kroymann, J. et al. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127, 1077–1088 (2001).
Kliebenstein, D.J., Lambrix, V.M., Reichelt, M., Gershenzon, J. & Mitchell-Olds, T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis . Plant Cell 13, 681–693 (2001).
de la Fuente, A., Bing, N., Hoeschele, I. & Mendes, P. Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics 20, 3565–3574 (2004).
Li, Y., Baldauf, S., Lim, E. & Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana . J. Biol. Chem. 276, 4338–4343 (2001).
Lim, E., Ashford, D.A., Hou, B., Jackson, G. & Bowles, D.J. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87, 623–631 (2004).
D'Auria, J.C. & Gershenzon, J. The secondary metabolism of Arabidopsis thaliana: growing like a weed. Curr. Opin. Plant Biol. 8, 308–316 (2005).
Alba, R. et al. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17, 2954–2965 (2005).
Lumba, S. & McCourt, P. Preventing leaf identity theft with hormones. Curr. Opin. Plant Biol. 8, 501–505 (2005).
Oksman-Caldentey, K.M. & Saito, K. Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 16, 174–179 (2005).
Lall, S., Nettleton, D., DeCook, R., Che, P. & Howell, S.H. Quantitative trait loci associated with adventitious shoot formation in tissue culture and the program of shoot development in Arabidopsis . Genetics 167, 1883–1892 (2004).
DeCook, R., Lall, S., Nettleton, D. & Howell, S.H. Genetic regulation of gene expression during shoot development in Arabidopsis . Genetics 172, 1155–1164 (2006).
Brem, R.B., Yvert, G., Clinton, R. & Kruglyak, L. Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755 (2002).
Schadt, E.E. et al. Genetics of gene expression surveyed in maize, mouse and man. Nature 422, 297–302 (2003).
Jansen, R.C. Studying complex biological systems using multifactorial perturbation. Nat. Rev. Genet. 4, 145–151 (2003).
Koornneef, M., Alonso-Blanco, C. & Vreugdenhil, D. Naturally occurring genetic variation in Arabidopsis thaliana . Annu. Rev. Plant Biol. 55, 141–172 (2004).
Stam, P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3, 739–744 (1993).
McCullagh, P. & Nelder, J.A. Generalized Linear Models (Chapman & Hall, New York, 1989).
Bing, N. & Hoeschele, I. Genetical genomics analysis of a yeast segregant population for transcription network inference. Genetics 170, 533–542 (2005).
Schadt, E.E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37, 710–717 (2005).
Zhu, J. et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet. Genome Res. 105, 363–374 (2004).
Acknowledgements
This work was supported by grants from the Netherlands Organization for Scientific Research, Program Genomics (050-10-029) and the Centre for Biosystems Genomics (CBSG, Netherlands Genomics Initiative).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Hierarchical clustering of accessions for metabolite content. (PDF 166 kb)
Supplementary Fig. 2
Frequency distribution of the number of QTLs detected for each mass signal. (PDF 3 kb)
Supplementary Fig. 3
Frequency distribution of broad sense heritability of each detected mass in the RIL population. (PDF 4 kb)
Supplementary Fig. 4
Frequency distribution of the contribution to QTL significance of the binominal part in the two-part parametric model. (PDF 294 kb)
Supplementary Fig. 5
Binominal and quantitative variance explained by QTLs. (PDF 558 kb)
Supplementary Table 1
Arabidopsis thaliana accessions used in the analysis of natural variation for metabolite content. (PDF 3 kb)
Supplementary Table 2
Detected QTL at and epistatic effects between MAM and AOP loci for aliphatic glucosinolates. (PDF 4 kb)
Supplementary Table 3
Phenotypic and mapping data of aliphatic glucosinolates. (PDF 6 kb)
Supplementary Table 4
Identification of flavonols. (PDF 5 kb)
Rights and permissions
About this article
Cite this article
Keurentjes, J., Fu, J., de Vos, C. et al. The genetics of plant metabolism. Nat Genet 38, 842–849 (2006). https://doi.org/10.1038/ng1815
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1815
This article is cited by
-
Metabolic-GWAS provides insights into genetic architecture of seed metabolome in buckwheat
BMC Plant Biology (2023)
-
Metabolomic profiling reveals shifts in defenses of an invasive plant
Biological Invasions (2023)
-
C. elegans as a model for inter-individual variation in metabolism
Nature (2022)
-
March of molecular breeding techniques in the genetic enhancement of herbal medicinal plants: present and future prospects
The Nucleus (2022)
-
OsRLCK160 contributes to flavonoid accumulation and UV-B tolerance by regulating OsbZIP48 in rice
Science China Life Sciences (2022)