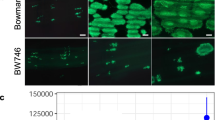
Abstract
In the fungal phylum Ascomycota, the ability to cause disease in plants and animals has been gained and lost repeatedly during phylogenesis1. In monocotyledonous barley, loss-of-function mlo alleles result in effective immunity against the Ascomycete Blumeria graminis f. sp. hordei, the causal agent of powdery mildew disease2,3. However, mlo-based disease resistance has been considered a barley-specific phenomenon to date. Here, we demonstrate a conserved requirement for MLO proteins in powdery mildew pathogenesis in the dicotyledonous plant species Arabidopsis thaliana. Epistasis analysis showed that mlo resistance in A. thaliana does not involve the signaling molecules ethylene, jasmonic acid or salicylic acid, but requires a syntaxin, glycosyl hydrolase and ABC transporter4,5,6. These findings imply that a common host cell entry mechanism of powdery mildew fungi evolved once and at least 200 million years ago, suggesting that within the Erysiphales (powdery mildews) the ability to cause disease has been a stable trait throughout phylogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Berbee, M.L. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant Pathol. 59, 165–187 (2001).
Jørgensen, J.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152 (1992).
Büschges, R. et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705 (1997).
Collins, N.C. et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977 (2003).
Lipka, V. et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183 (2005).
Stein, M. et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746 (2006).
Piffanelli, P. et al. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891 (2004).
Panstruga, R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33, 389–392 (2005).
Devoto, A. et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56, 77–88 (2003).
Panstruga, R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol. Biol. 59, 485–500 (2005).
Vogel, J. & Somerville, S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97, 1897–1902 (2000).
Vogel, J.P., Raab, T.K., Schiff, C. & Somerville, S.C. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106 (2002).
Vogel, J.P., Raab, T.K., Somerville, C.R. & Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40, 968–978 (2004).
Jarosch, B., Kogel, K.H. & Schaffrath, U. The ambivalence of the barley Mlo locus: Mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 12, 508–514 (1999).
Kumar, J., Hückelhoven, R., Beckhove, U., Nagarajan, S. & Kogel, K.H. A compromised Mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (Teleomorph: Cochliobolus sativus) and its toxins. Phytopathology 91, 127–133 (2001).
Wolter, M., Hollricher, K., Salamini, F. & Schulze-Lefert, P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defense mimic phenotype. Mol. Gen. Genet. 239, 122–128 (1993).
Peterhänsel, C., Freialdenhoven, A., Kurth, J., Kolsch, R. & Schulze-Lefert, P. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell 9, 1397–1409 (1997).
Piffanelli, P. et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085 (2002).
Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis - 2001 status. Curr. Opin. Plant Biol. 4, 301–308 (2001).
van Wees, S.C. & Glazebrook, J. Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742 (2003).
Morris, K. et al. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23, 677–685 (2000).
Dong, X.N. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323 (1998).
Freialdenhoven, A., Peterhänsel, C., Kurth, J., Kreuzaler, F. & Schulze-Lefert, P. Identification of genes required for the function of non-race- specific mlo resistance to powdery mildew in barley. Plant Cell 8, 5–14 (1996).
Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P. & Panstruga, R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102, 3135–3140 (2005).
Xu, Z. et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367 (2004).
Wolfe, K.H., Gouy, M.L., Yang, Y.W., Sharp, P.M. & Li, W.H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205 (1989).
Mori, Y., Sato, Y. & Takamatsu, S. Molecular phylogeny and radiation time of Erysiphales inferred from the nuclear ribosomal DNA sequences. Mycoscience 41, 437–447 (2000).
Galagan, J.E. et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115 (2005).
Acknowledgements
We thank T. Gjetting (Biotechnology Research and Innovation Centre, Denmark) and J. Shrager (Carnegie Institution) for helpful discussions. We acknowledge the Salk Institute, The Sainsbury Laboratory, Syngenta and the Arabidopsis Biological Resource Center for providing T-DNA/transposon insertion lines and some of the mutant lines. This work was supported by grants from the Max-Planck Society and the Deutsche Forschungsgemeinschaft (PA861/4; to R.P.); grants from the US National Science Foundation (0114783 and 0519898) and the Carnegie Institution (to S.C.S.) and a US National Institutes of Health (NIH) fellowship (FG32 GN19499-01 to J.V.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
AtMLO co-orthologs are required for susceptibility to two powdery mildew species. (PDF 237 kb)
Supplementary Fig. 2
Interactions between Atmlo mutants and nonadapted pathogens. (PDF 74 kb)
Supplementary Fig. 3
Atmlo mutants exhibit constitutively elevated levels of salicylic acid. (PDF 23 kb)
Supplementary Fig. 4
Enhanced powdery mildew host cell entry in Atmlo2 NahG plants is likely due to non-SA-dependent defense responses. (PDF 21 kb)
Supplementary Fig. 5
The PEN2 glycosyl hydrolase is an evolutionarily recent acquisition. (PDF 24 kb)
Supplementary Table 1
Atmlo alleles used in this study. (PDF 48 kb)
Supplementary Table 2
Nucleotide sequences of oligonucleotides used in this study. (PDF 7 kb)
Rights and permissions
About this article
Cite this article
Consonni, C., Humphry, M., Hartmann, H. et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38, 716–720 (2006). https://doi.org/10.1038/ng1806
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1806
This article is cited by
-
Application of bulk segregant RNA-Seq (BSR-Seq) and allele-specific primers to study soybean powdery mildew resistance
BMC Plant Biology (2024)
-
An Overview of Targeted Genome Editing Strategies for Reducing the Biosynthesis of Phytic Acid: an Anti-nutrient in Crop Plants
Molecular Biotechnology (2024)
-
Functional and evolutionary study of MLO gene family in the regulation of Sclerotinia stem rot resistance in Brassica napus L.
Biotechnology for Biofuels and Bioproducts (2023)
-
Enhancing powdery mildew resistance in soybean by targeted mutation of MLO genes using the CRISPR/Cas9 system
BMC Plant Biology (2023)
-
RALF signaling pathway activates MLO calcium channels to maintain pollen tube integrity
Cell Research (2023)