Abstract
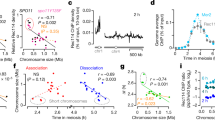
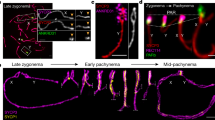
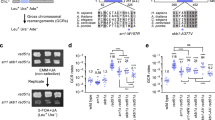
Errors in meiotic chromosome segregation are the leading cause of spontaneous abortions and birth defects1. In humans, chromosomes that fail to experience crossovers (or exchanges) are error-prone, more likely than exchange chromosomes to mis-segregate in meiosis. We used a yeast model to investigate the mechanisms that partition nonexchange chromosomes. These studies showed that the spindle checkpoint genes MAD1, MAD2 and MAD3 have different roles. We identified a new meiotic role for MAD3; though dispensable for the segregation of exchange chromosomes, it is essential for the segregation of nonexchange chromosomes. This function of Mad3p could also be carried out by human BubR1. MAD1 and MAD2 act in a surveillance mechanism that mediates a metaphase delay in response to nonexchange chromosomes, whereas MAD3 acts as a crucial meiotic timer, mediating a prophase delay in every meiosis. These findings suggest plausible models for the basis of errant meiotic segregation in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001).
Hunt, P.A. & Hassold, T.J. Sex matters in meiosis. Science 296, 2181–2183 (2002).
Baudat, F., Manova, K., Yuen, J.P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Lamb, N.E. et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14, 400–405 (1996).
Volarcik, K. et al. The meiotic competence of in-vitro matured human oocytes is influenced by donor age: evidence that folliculogenesis is compromised in the reproductively aged ovary. Hum. Reprod. 13, 154–160 (1998).
Steuerwald, N., Cohen, J., Herrera, R.J., Sandalinas, M. & Brenner, C.A. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol. Hum. Reprod. 7, 49–55 (2001).
Hodges, C.A. et al. Experimental evidence that changes in oocyte growth influence meiotic chromosome segregation. Hum. Reprod. 17, 1171–1180 (2002).
Kong, A. et al. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206 (2004).
Boumil, R.M., Kemp, B., Angelichio, M., Nilsson-Tillgren, T. & Dawson, D.S. Meiotic segregation of a homeologous chromosome pair. Mol. Genet. Genomics 268, 750–760 (2003).
Kaback, D.B., Guacci, V., Barber, D. & Mahon, J.W. Chromosome size-dependent control of meiotic recombination. Science 256, 228–232 (1992).
Li, R. & Murray, A.W. Feedback control of mitosis in budding yeast. Cell 66, 519–531 (1991).
Shonn, M.A., McCarroll, R. & Murray, A.W. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289, 300–303 (2000).
Shonn, M.A., Murray, A.L. & Murray, A.W. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr. Biol. 13, 1979–1984 (2003).
Lew, D.J. & Burke, D.J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37, 251–282 (2003).
Chan, G.K., Jablonski, S.A., Sudakin, V., Hittle, J.C. & Yen, T.J. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146, 941–954 (1999).
Meraldi, P., Draviam, V.M. & Sorger, P.K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45–60 (2004).
Kemp, B., Boumil, R.M., Stewart, M.N. & Dawson, D.S. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 18, 1946–1951 (2004).
Nicklas, R.B. & Koch, C.A. Chromosome manipulation III. Spindle fiber tension and the re-orientation of mal-oriented chromosomes. J. Cell Biol. 43, 40–50 (1969).
Nicklas, R.B. How cells get the right chromosomes. Science 275, 632–637 (1997).
Wells, W.A. & Murray, A.W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J. Cell Biol. 133, 75–84 (1996).
Wassmann, K., Niault, T. & Maro, B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr. Biol. 13, 1596–1608 (2003).
Zhang, D. et al. Localization of mitotic arrest deficient 1 (MAD1) in mouse oocytes during the first meiosis and its functions as a spindle checkpoint protein. Biol. Reprod. 72, 58–68 (2005).
Homer, H.A. et al. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 19, 202–207 (2005).
Gillett, E.S., Espelin, C.W. & Sorger, P.K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164, 535–546 (2004).
Brunet, S. et al. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J. Cell Biol. 146, 1–12 (1999).
Baker, D.J. et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36, 744–749 (2004).
Burke, D., Dawson, D. & Stearns, T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2000).
Nicolas, A., Treco, D., Schultes, N.P. & Szostak, J.W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae . Nature 338, 35–39 (1989).
Longtine, M.S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14, 953–961 (1998).
Haase, S.B. & Reed, S.I. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1, 132–136 (2002).
Acknowledgements
We thank M. Shonn Dorer, B. McCarroll, A. Murray and A. Straight for providing key reagents and for many discussions of this work; M. Stewart for critical reading of the manuscript; members of the laboratory of D.S.D. for their contributions to the project; T. Stearns and M. Winey for antibodies to Tub4; F. McKeon for hBubR1 clones; and S. Acharya and A. Parmalee for assistance with flow cytometry. This research was supported by a grant from the March of Dimes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Strains used in this study. (PDF 106 kb)
Rights and permissions
About this article
Cite this article
Cheslock, P., Kemp, B., Boumil, R. et al. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat Genet 37, 756–760 (2005). https://doi.org/10.1038/ng1588
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1588
This article is cited by
-
Inter-telomeric connections and achiasmate meiosis in Tradescantia spathacea Sw.
The Nucleus (2016)
-
Couples, pairs, and clusters: mechanisms and implications of centromere associations in meiosis
Chromosoma (2014)
-
Finding a match: how do homologous sequences get together for recombination?
Nature Reviews Genetics (2008)
-
The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere
Nature Genetics (2007)
-
A delay like no other
Nature Genetics (2005)