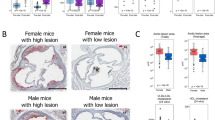
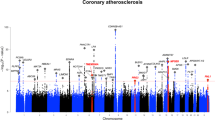
Abstract
Ath1 is a quantitative trait locus on mouse chromosome 1 that renders C57BL/6 mice susceptible and C3H/He mice resistant to diet-induced atherosclerosis. The quantitative trait locus region encompasses 11 known genes, including Tnfsf4 (also called Ox40l or Cd134l), which encodes OX40 ligand. Here we report that mice with targeted mutations of Tnfsf4 had significantly (P ≤ 0.05) smaller atherosclerotic lesions than did control mice. In addition, mice overexpressing Tnfsf4 had significantly (P ≤ 0.05) larger atherosclerotic lesions than did control mice. In two independent human populations, the less common allele of SNP rs3850641 in TNFSF4 was significantly more frequent (P ≤ 0.05) in individuals with myocardial infarction than in controls. We therefore conclude that Tnfsf4 underlies Ath1 in mice and that polymorphisms in its human homolog TNFSF4 increase the risk of myocardial infarction in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Paigen, B., Morrow, A., Brandon, C., Mitchell, D. & Holmes, P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 57, 65–73 (1985).
Paigen, B. et al. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc. Natl. Acad. Sci. USA 84, 3763–3767 (1987).
Paigen, B., Albee, D., Holmes, P.A. & Mitchell, D. Genetic analysis of murine strains C57BL/6J and C3H/HeJ to confirm the map position of Ath-1, a gene determining atherosclerosis susceptibility. Biochem. Genet. 25, 501–511 (1987).
Phelan, S.A., Beier, D.R., Higgins, D.C. & Paigen, B. Confirmation and high resolution mapping of an atherosclerosis susceptibility gene in mice on Chromosome 1. Mamm. Genome 13, 548–553 (2002).
Wang, X. et al. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J. Biol. Chem. 278, 25179–25190 (2003).
Phelan, S.A., Wang, X., Wallbrandt, P., Forsman-Semb, K. & Paigen, B. Overexpression of Prdx6 reduces H2O2 but does not prevent diet-induced atherosclerosis in the aortic root. Free Radic. Biol. Med. 35, 1110–1120 (2003).
Wang, X. et al. Peroxiredoxin 6 deficiency and atherosclerosis susceptibility in mice: significance of genetic background for assessing atherosclerosis. Atherosclerosis 177, 61–70 (2004).
Geng, Y.J. Biologic effect and molecular regulation of vascular apoptosis in atherosclerosis. Curr. Atheroscler. Rep. 3, 234–242 (2001).
Weinberg, A.D. OX40: targeted immunotherapy–implications for tempering autoimmunity and enhancing vaccines. Trends Immunol. 23, 102–109 (2002).
Hansson, G.K., Libby, P., Schonbeck, U. & Yan, Z.Q. Innate and adaptive immunity in the pathogenesis of atherosclerosis. Circ. Res. 91, 281–291 (2002).
Shi, W. et al. Genetic backgrounds but not sizes of atherosclerotic lesions determine medial destruction in the aortic root of apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 23, 1901–1906 (2003).
Mehrabian, M. et al. Identification of 5-lipoxygenase as a major gene contributing to atherosclerosis susceptibility in mice. Circ. Res. 91, 120–126 (2002).
Emeson, E.E., Shen, M.L., Bell, C.G. & Qureshi, A. Inhibition of atherosclerosis in CD4 T-cell-ablated and nude (nu/nu) C57BL/6 hyperlipidemic mice. Am. J. Pathol. 149, 675–685 (1996).
Nakai, Y. et al. Natural killer T cells accelerate atherogenesis in mice. Blood 104, 2051–2059 (2004).
Major, A.S., Fazio, S. & Linton, M.F. B-lymphocyte deficiency increases atherosclerosis in LDL receptor-null mice. Arterioscler. Thromb. Vasc. Biol. 22, 1892–1898 (2002).
Caligiuri, G., Nicoletti, A., Poirier, B. & Hansson, G.K. Protective immunity against atherosclerosis carried by B cells of hypercholesterolemic mice. J. Clin. Invest. 109, 745–753 (2002).
Reardon, C.A. et al. Effect of immune deficiency on lipoproteins and atherosclerosis in male apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 21, 1011–1016 (2001).
Song, L., Leung, C. & Schindler, C. Lymphocytes are important in early atherosclerosis. J. Clin. Invest. 108, 251–259 (2001).
Dansky, H.M., Charlton, S.A., Harper, M.M. & Smith, J.D. T and B lymphocytes play a minor role in atherosclerotic plaque formation in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. USA 94, 4642–4646 (1997).
Geng, Y.J. & Libby, P. Progression of atheroma: a struggle between death and procreation. Arterioscler. Thromb. Vasc. Biol. 22, 1370–1380 (2002).
Yang, J. et al. Endothelial overexpression of Fas ligand decreases atherosclerosis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 24, 1466–1473 (2004).
Aprahamian, T. et al. Impaired clearance of apoptotic cells promotes synergy between atherogenesis and autoimmune disease. J. Exp. Med. 199, 1121–1131 (2004).
Schneider, D.B. et al. Expression of Fas ligand in arteries of hypercholesterolemic rabbits accelerates atherosclerotic lesion formation. Arterioscler. Thromb. Vasc. Biol. 20, 298–308 (2000).
Kotani, A., Hori, T., Matsumura, Y. & Uchiyama, T. Signaling of gp34 (OX40 ligand) induces vascular endothelial cells to produce a CC chemokine RANTES/CCL5. Immunol. Lett. 84, 1–7 (2002).
Kawai, T. et al. Selective diapedesis of Th1 cells induced by endothelial cell RANTES. J. Immunol. 163, 3269–3278 (1999).
Welch, C.L. et al. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc. Natl. Acad. Sci. USA 98, 7946–7951 (2001).
Hauser, E.R. et al. A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am. J. Hum. Genet. 75, 436–447 (2004).
Wang, Q. et al. Premature myocardial infarction novel susceptibility locus on chromosome 1p34-36 identified by genomewide linkage analysis. Am. J. Hum. Genet. 74, 262–271 (2004).
Abiola, O. et al. The nature and identification of quantitative trait loci: a community's view. Nat. Rev. Genet. 4, 911–916 (2003).
Stary, H.C. et al. A definition of advanced types of atherosclerotic lesions and a histological classification of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation 92, 1355–1374 (1995).
Edfeldt, K. et al. Association of hypo-responsive toll-like receptor 4 variants with risk of myocardial infarction. Eur. Heart J. 25, 1447–1453 (2004).
Paigen, B., Holmes, P.A., Mitchell, D. & Albee, D. Comparison of atherosclerotic lesions and HDL-lipid levels in male, female, and testosterone-treated female mice from strains C57BL/6, BALB/c, and C3H. Atherosclerosis 64, 215–221 (1987).
Sugamura, K., Ishii, N. & Weinberg, A.D. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat. Rev. Immunol. 4, 420–431 (2004).
Murata, K. et al. Impairment of antigen-presenting cell function in mice lacking expression of OX40 ligand. J. Exp. Med. 191, 365–374 (2000).
Nishina, P.M., Verstuyft, J. & Paigen, B. Synthetic low and high fat diets for the study of atherosclerosis in the mouse. J. Lipid Res. 31, 859–869 (1990).
Paigen, B. et al. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc. Natl. Acad. Sci. USA 84, 3763–3767 (1987).
Paigen, B., Mitchell, D., Holmes, P.A. & Albee, D. Genetic analysis of strains C57BL/6J and BALB/cJ for Ath-1, a gene determining atherosclerosis susceptibility in mice. Biochem. Genet. 25, 881–892 (1987).
Paigen, B., Albee, D., Holmes, P.A. & Mitchell, D. Genetic analysis of murine strains C57BL/6J and C3H/HeJ to confirm the map position of Ath-1, a gene determining atherosclerosis susceptibility. Biochem. Genet. 25, 501–511 (1987).
Paigen, B., Morrow, A., Holmes, P.A., Mitchell, D. & Williams, R.A. Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis 68, 231–240 (1987).
Wang, X. et al. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome. Res. 13, 1654–1664 (2003).
Eriksson, P. et al. Human evidence that the cystatin C gene is implicated in focal progression of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 24, 551–557 (2004).
Reuterwall, C. et al. Higher relative, but lower absolute risks of myocardial infarction in women than in men: analysis of some major risk factors in the SHEEP study. The SHEEP Study Group. J. Intern. Med. 246, 161–174 (1999).
Boquist, S. et al. Alimentary lipemia, postprandial triglyceride-rich lipoproteins, and common carotid intima-media thickness in healthy, middle-aged men. Circulation 100, 723–728 (1999).
Austen, W.G. et al. A reporting system on patients evaluated for coronary artery disease. Report of the Ad Hoc Committee for Grading of Coronary Artery Disease, Council on Cardiovascular Surgery, American Heart Association. Circulation 51, 5–40 (1975).
Ronaghi, M., Uhlen, M. & Nyren, P. A sequencing method based on real-time pyrophosphate. Science 281, 363–365 (1998).
Niu, T., Qin, Z.S., Xu, X. & Liu, J.S. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am. J. Hum. Genet. 70, 157–169 (2002).
Acknowledgements
We thank C. McFarland for technical assistance and R. Lambert for helping to prepare the manuscript. This study was supported by AstraZeneca (Sweden), grants from the US National Institutes of Health and grants from the Swedish Heart-Lung Foundation, the Swedish Medical Research Council, the Torsten and Ragnar Söderberg foundation, AFA insurance, the Stockholm County Council, the Wallenberg Consortium North, the Swedish Society for Medical Research and the Karolinska Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Differences in 5′ upstream sequences between B6 and C3H Ox40l. (PDF 22 kb)
Supplementary Table 2
Allele frequencies and pairwise linkage disequilibrium coefficients for the SNPs in the OX40L gene in control subjects. (PDF 19 kb)
Supplementary Table 3
Association of the 110NN haplotype with the levels of plasma lipids and serum amyloid A in control subjects. (PDF 20 kb)
Supplementary Table 4
Primers for genotyping, RT-PCR and sequencing. (PDF 21 kb)
Supplementary Table 5
OX40L sequencing primers (PDF 17 kb)
Supplementary Table 6
Pyrosequencing© primers for OX40L (PDF 18 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Ria, M., Kelmenson, P. et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet 37, 365–372 (2005). https://doi.org/10.1038/ng1524
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1524
This article is cited by
-
Genetic susceptibility to diabetic kidney disease is linked to promoter variants of XOR
Nature Metabolism (2023)
-
Genetics of murine type 2 diabetes and comorbidities
Mammalian Genome (2022)
-
Phenotypic characteristics of commonly used inbred mouse strains
Journal of Molecular Medicine (2020)
-
The role of OX40L and ICAM-1 in the stability of coronary atherosclerotic plaques and their relationship with sudden coronary death
BMC Cardiovascular Disorders (2019)
-
Engineering Nanomaterials to Address Cell-Mediated Inflammation in Atherosclerosis
Regenerative Engineering and Translational Medicine (2016)