Abstract
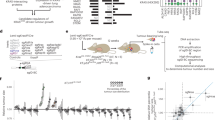
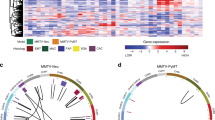
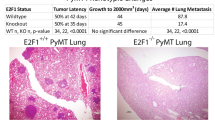
Using advanced gene targeting methods, generating mouse models of cancer that accurately reproduce the genetic alterations present in human tumors is now relatively straightforward. The challenge is to determine to what extent such models faithfully mimic human disease with respect to the underlying molecular mechanisms that accompany tumor progression. Here we describe a method for comparing mouse models of cancer with human tumors using gene-expression profiling. We applied this method to the analysis of a model of Kras2-mediated lung cancer and found a good relationship to human lung adenocarcinoma, thereby validating the model. Furthermore, we found that whereas a gene-expression signature of KRAS2 activation was not identifiable when analyzing human tumors with known KRAS2 mutation status alone, integrating mouse and human data uncovered a gene-expression signature of KRAS2 mutation in human lung cancer. We confirmed the importance of this signature by gene-expression analysis of short hairpin RNA–mediated inhibition of oncogenic Kras2. These experiments identified both a pattern of gene expression indicative of KRAS2 mutation and potential effectors of oncogenic KRAS2 activity in human cancer. This approach provides a strategy for using genomic analysis of animal models to probe human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
van 't Veer, L.J. et al. Expression profiling predicts outcome in breast cancer. Breast Cancer Res 5, 57–58 (2002).
Ramaswamy, S., Ross, K.N., Lander, E.S. & Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 (2003).
Chang, H.Y. et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2, E7 (2004).
Armstrong, S.A. et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30, 41–47 (2002).
Ferrando, A.A. et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1, 75–87 (2002).
Ross, M.E. et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 102, 2951–2959 (2003).
Desai, K.V. et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc. Natl. Acad. Sci. USA 99, 6967–6972 (2002).
Huang, E. et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat. Genet. 34, 226–230 (2003).
Tuveson, D.A. et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5, 375–387 (2004).
Repasky, G.A., Chenette, E.J. & Der, C.J. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 14, 639–647 (2004).
Zuber, J. et al. A genome-wide survey of RAS transformation targets. Nat. Genet. 24, 144–152 (2000).
Tchernitsa, O.I. et al. Transcriptional basis of KRAS oncogene-mediated cellular transformation in ovarian epithelial cells. Oncogene 23, 4536–4555 (2004).
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Bhattacharjee, A. et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA 98, 13790–13795 (2001).
Vapnik, V.N. Statistical Learning Theory (John Wiley and Sons, New York, 1998).
Ramaswamy, S. et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. USA 98, 15149–15154 (2001).
Lamb, J. et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114, 323–334 (2003).
Mootha, V.K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Bonner, A.E., Lemon, W.J., Devereux, T.R., Lubet, R.A. & You, M. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene 23, 1166–1176 (2004).
Lo, R.S., Wotton, D. & Massague, J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 20, 128–136 (2001).
Beer, D.G. et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8, 816–824 (2002).
Naoki, K., Chen, T.H., Richards, W.G., Sugarbaker, D.J. & Meyerson, M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 62, 7001–7003 (2002).
Paez, J.G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Iacobuzio-Donahue, C.A. et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 162, 1151–1162 (2003).
Klimstra, D.S. & Longnecker, D.S. K-ras mutations in pancreatic ductal proliferative lesions. Am. J. Pathol. 145, 1547–1550 (1994).
Peeper, D.S. et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386, 177–181 (1997).
Camps, M. et al. Induction of the mitogen-activated protein kinase phosphatase MKP3 by nerve growth factor in differentiating PC12. FEBS Lett. 425, 271–276 (1998).
Muda, M. et al. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J. Biol. Chem. 271, 27205–27208 (1996).
Kawakami, Y. et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 5, 513–519 (2003).
Serrano, M., Lin, A.W., McCurrach, M.E., Beach, D. & Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Toyoshima, Y. et al. TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J. Biol. Chem. 279, 25898–25904 (2004).
Ellwood-Yen, K. et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003).
Golub, T.R. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537 (1999).
Rubinson, D.A. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33, 401–406 (2003).
Mantel, N. The detection of disease clustering and a generalized regression approach 27, 209–220 (1967).
Acknowledgements
We thank P. Tamayo and K. Haigis for comments and critical review of the manuscript and M. You for providing access to the gene expression data and histology slides for the NNK mouse models. This work was supported in part by the National Institutes of Health and the National Cancer Institute. T.J. and T.R.G. are investigators of the Howard Hughes Medical Institute. A.S.-C. was supported in part by grants from the Robert Woods Johnson Foundation (Harold Amos Medical Faculty Development Program) and by a mentored clinical scientist grant from the National Cancer Institute. S.M. received partial support from an Alfred P. Sloan Foundation/U.S. Department of Energy Fellowship in Computational Molecular Biology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Representative example of KrasLA murine lung tumor histology. (PDF 40 kb)
Supplementary Fig. 2
Representative example of Bonner et al carcinogen induced lung tumor histology. (PDF 70 kb)
Supplementary Fig. 3
Distribution of ES scores generated from random gene sets. (PDF 171 kb)
Supplementary Fig. 4
ES scores obtained from gene sets from permuted mouse phenotypes. (PDF 171 kb)
Supplementary Table 1
KrasLA model up-regulated gene set. (XLS 152 kb)
Supplementary Table 2
KrasLA model down-regulated gene set. (XLS 130 kb)
Supplementary Table 3
Venn diagram of KrasLA model and various cancer subtypes. (PDF 51 kb)
Supplementary Table 4
Adenocarcinoma signature. (XLS 42 kb)
Supplementary Table 5
GSEA results for Boston & Ann Arbor lung adenocarcinomas Kras mutant vs. wild-type. (PDF 44 kb)
Supplementary Table 6
Kras signature. (XLS 39 kb)
Supplementary Table 7
GSEA of Kras signature on human datasets. (PDF 46 kb)
Supplementary Table 8
Kras signature in A549 knockdown. (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Sweet-Cordero, A., Mukherjee, S., Subramanian, A. et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 37, 48–55 (2005). https://doi.org/10.1038/ng1490
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1490
This article is cited by
-
RAS oncogenic activity predicts response to chemotherapy and outcome in lung adenocarcinoma
Nature Communications (2022)
-
KRAS drives immune evasion in a genetic model of pancreatic cancer
Nature Communications (2021)
-
Enhancing chemotherapy response through augmented synthetic lethality by co-targeting nucleotide excision repair and cell-cycle checkpoints
Nature Communications (2020)
-
An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer
Nature Communications (2017)
-
Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma
Nature Medicine (2016)