Abstract
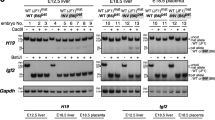
Imprinted expression at the H19-Igf2 locus depends on a differentially methylated domain (DMD) that acts both as a maternal-specific, methylation-sensitive insulator and as a paternal-specific site of hypermethylation. Four repeats in the DMD bind CCCTC-binding factor (CTCF) on the maternal allele and have been proposed to attract methylation on the paternal allele. We introduced point mutations into the DMD to deplete the repeats of CpGs while retaining CTCF-binding and enhancer-blocking activity. Maternal inheritance of the mutations left H19 expression and Igf2 imprinting intact, consistent with the idea that the DMD acts as an insulator. Conversely, paternal inheritance of these mutations disrupted maintenance of DMD methylation, resulting in biallelic H19 expression. Furthermore, an insulator was established on the paternally inherited mutated allele in vivo, reducing Igf2 expression and resulting in a 40% reduction in size of newborn offspring. Thus, the nine CpG mutations in the DMD showed that the two parental-specific roles of the H19 DMD, methylation maintenance and insulator assembly, are antagonistic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Verona, R.I., Mann, M.R.W. & Bartolomei, M.S. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell. Dev. Biol. 19, 237–258 (2003).
Thorvaldsen, J.L., Mann, M.R., Nwoko, O., Duran, K.L. & Bartolomei, M.S. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22, 2450–2462 (2002).
Lopes, S. et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 12, 295–305 (2003).
Eden, S. et al. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 20, 3518–3525 (2001).
Ainscough, J.F.-X., Dandolo, L. & Surani, M.A. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech. Dev. 91, 365–368 (2000).
Leighton, P.A., Saam, J.R., Ingram, R.S., Stewart, C.L. & Tilghman, S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9, 2079–2089 (1995).
Thorvaldsen, J.L., Duran, K.L. & Bartolomei, M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12, 3693–3702 (1998).
DeChiara, T.M., Efstratiadis, A. & Robertson, E.J. A growth-deficiency phenotype in heterozygous mice carrying an insulin- like growth factor II gene disrupted by targeting. Nature 345, 78–80 (1990).
Tremblay, K.D., Duran, K.L. & Bartolomei, M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17, 4322–4329 (1997).
Davis, T.L., Trasler, J.M., Moss, S.B., Yang, G.J. & Bartolomei, M.S. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58, 18–28 (1999).
Bartolomei, M.S., Webber, A.L., Brunkow, M.E. & Tilghman, S.M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7, 1663–1673 (1993).
Ferguson-Smith, A.S., Sasaki, H., Cattanach, B.M. & Surani, M.A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–754 (1993).
Li, E., Beard, C. & Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993).
Stadnick, M.P. et al. Role of a 461 bp G-rich repetitive element in H19 transgene imprinting. Dev. Genes Evol. 209, 239–248 (1999).
Hark, A.T. et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000).
Bell, A.C. & Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 (2000).
Frevel, M.A.E., Hornberg, J.J. & Reeve, A.E. A potential imprint control element: identification of a conserved 42 bp sequence upstream of H19. Trends Genet. 15, 216–218 (1999).
Bell, A.C., West, A.G. & Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 (1999).
Srivastava, M. et al. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 14, 1186–1195 (2000).
Mann, M.R. et al. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69, 902–914 (2003).
Kanduri, C. et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation sensitive. Curr. Biol. 10, 853–856 (2000).
Schoenherr, C.J., Levorse, J.M. & Tilghman, S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33, 66–69 (2003).
Pant, V. et al. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17, 586–590 (2003).
Neumann, B., Kubicka, P. & Barlow, D.P. Characteristics of imprinted genes. Nat. Genet. 9, 12–13 (1995).
Yoon, B.J. et al. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30, 92–96 (2002).
Fedoriw, A.M., Stein, P., Svoboda, P., Schultz, R.M. & Bartolomei, M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240 (2004).
Lewandoski, M., Wassarman, K.M. & Martin, G.R. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148–151 (1997).
Auffray, C. & Rougeon, F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur. J. Biochem. 107, 303–314 (1980).
Chung, J.H., Whiteley, M. & Felsenfeld, G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74, 505–514 (1993).
Chung, J.H., Bell, A.C. & Felsenfeld, G. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. USA 94, 575–580 (1997).
Acknowledgements
We thank J. Richa at the University of Pennsylvania Transgenic Core Facility for the chimeric mice; S. Lee, C. Krapp and J. Piel for technical assistance; J. Thorvaldsen, R. Verona and J. Mager for critical reading of the manuscript; and M. Donohoe for advice with the ChIP assay. This work was supported by a US National Institutes of Health grant and the Howard Hughes Medical Institute. N.E. is the recipient of a National Research Service Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Engel, N., West, A., Felsenfeld, G. et al. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet 36, 883–888 (2004). https://doi.org/10.1038/ng1399
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1399
This article is cited by
-
Loss of SRSF2 triggers hepatic progenitor cell activation and tumor development in mice
Communications Biology (2020)
-
YY1’s role in the Peg3 imprinted domain
Scientific Reports (2017)
-
CTCF participates in DNA damage response via poly(ADP-ribosyl)ation
Scientific Reports (2017)
-
Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations
Clinical Epigenetics (2016)
-
Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines
BMC Cancer (2016)