Abstract
Inflammation influences iron balance in the whole organism. A common clinical manifestation of these changes is anemia of chronic disease (ACD; also called anemia of inflammation). Inflammation reduces duodenal iron absorption and increases macrophage iron retention, resulting in low serum iron concentrations (hyposideremia). Despite the protection hyposideremia provides against proliferating microorganisms, this 'iron withholding' reduces the iron available to maturing red blood cells and eventually contributes to the development of anemia1. Hepcidin antimicrobial peptide (Hamp) is a hepatic defensin-like peptide hormone2,3 that inhibits duodenal iron absorption and macrophage iron release4,5,6,7. Hamp is part of the type II acute phase response8 and is thought to have a crucial regulatory role in sequestering iron in the context of ACD7,9. Mice with deficiencies in the hemochromatosis gene product, Hfe, mounted a general inflammatory response after injection of lipopolysaccharide but lacked appropriate Hamp expression and did not develop hyposideremia. These data suggest a previously unidentified role for Hfe in innate immunity and ACD.
Similar content being viewed by others
Main
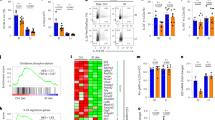
Hamp expression is induced in response to iron overload and inflammation4,6. Hfe is known to be required for this induction in response to iron overload10,11, and we tested whether it is also required for induction of Hamp in response to inflammation. We used Hfe-deficient mice homozygous with respect to either a null allele of Hfe (Hfe−/− ) or the 882G→A mutation (Hfe882A/882A ), which is orthologous to the hereditary hemochromatosis founder mutation in humans (845G→A in HFE)12. We treated 8-week-old female 129S6/SvEvTac wild-type or Hfe-deficient mice with 5 μg of lipopolysaccharide (LPS) to elicit the acute phase response and then killed the mice after 1.5 h. Northern-blot analysis of liver total RNA confirmed that Hamp expression was induced by a factor of 1.8 in wild-type mice treated with LPS (Fig. 1). In Hfe-deficient mice (Fig. 1), Hamp expression was not substantially induced in response to LPS, suggesting that Hfe is required to mediate an appropriate Hamp response to inflammation.
We used northern blotting to analyze Hamp expression in the livers of 8-week-old female mice that were untreated or treated with LPS (n = 4 mice in each group), compared the amounts of Hamp and Gapd mRNA and plotted this ratio. Bars represent s.d. Hamp expression was 1.8 times higher in wild-type (WT) mice but not in Hfe-deficient mice 1.5 h after treatment with LPS.
Tumor necrosis factor-α (Tnf) and Interleukin-6 (Il6) are proinflammatory cytokines that are induced in response to LPS. They trigger the expression of acute phase genes13. Il6 induces Hamp expression in isolated hepatocytes8. To determine whether the type I and type II acute phase responses14,15 were intact in Hfe−/− mice, we assayed Tnf and Il6 expression by enzyme-linked immunosorbent assays (ELISAs) in 8-week-old female mice either left untreated or treated with LPS and killed after 1.5 h (Fig. 2a,b). As in wild-type mice, Tnf expression was more than 275 times higher (Fig. 2a) and Il6 expression was more than 775 times higher (Fig. 2b) in Hfe−/− mice in response to LPS. We confirmed this induction of Tnf and Il6 by quantitative RT-PCR (Fig. 2c,d). These data suggest that LPS is equally efficient at cytokine induction in Hfe-deficient and wild-type mice. Therefore, the lack of appropriate Hamp expression in Hfe-deficient mice is not due to an insufficient acute phase response. More importantly, these data indicate that Hfe acts downstream, or independent of, Il6 in the inflammatory cascade that induces Hamp expression.
We assayed cytokine expression in the serum of 8-week-old female mice that were untreated or treated with LPS and killed after 1.5 h. WT, wild-type. In each panel, bars represent s.d. (a) Because Tnf expression in untreated mice was less than the lowest standard of 23.4 pg ml−1, we used the concentration of the lowest standard to calculate significance of differences between untreated and LPS-treated mice (n = 4 in each group). Tnf levels increased to 7,673.4 pg ml−1 in wild-type mice treated with LPS and to 6,473.0 pg ml−1 in Hfe−/− mice treated with LPS. (b) Il6 levels increased from 39.4 pg ml−1 to 30,823.9 pg ml−1 in wild-type mice treated with LPS and from 41.2 pg ml−1 to 29,062.8 pg ml−1 in Hfe−/− mice treated with LPS. n = 4 in each group. (c) We further assessed Tnf mRNA expression by quantitative RT-PCR. Tnf mRNA expression increased by a factor of 30.5 in wild-type mice treated with LPS, by a factor of 82.3 in Hfe−/− mice treated with LPS and by a factor of 99.1 in mutant Hfe882A/882A mice treated with LPS. n = 4–6 in each group. (d) We further assessed Il6 mRNA expression by quantitative RT-PCR. Il6 mRNA expression increased by a factor of 7.1 in wild-type mice treated with LPS, by a factor of 8.9 in Hfe−/− mice treated with LPS and by a factor of 13.3 in Hfe882A/882A mice treated with LPS. n = 4–6 in each group.
LPS-induced inflammation mimics ACD in that it induces hyposideremia in mice16. To assay for hyposideremia, we analyzed serum iron concentrations in 8-week-old female mice treated with LPS. Serum iron concentrations were not substantially lower in wild-type or Hfe-deficient mice 1.5 h after LPS treatment (data not shown). But 24 h after treatment with LPS, serum iron concentrations in wild-type mice dropped significantly from 185 mg dl−1 to 99 mg dl−1 (Table 1). Il6 and Tnf levels returned to baseline 24 h after treatment in wild-type mice, and Hamp expression was no longer increased (data not shown). Transferrin (Trf) saturation decreased from 68% to 39% in wild-type mice 24 h after treatment with LPS (Table 1), owing to the decrease in serum iron concentration; accordingly, the total iron binding capacity did not change significantly (Table 1). In contrast to wild-type controls, Hfe−/− mice did not develop hyposideremia in response to LPS (Table 1). Hfe−/− mice had higher Trf saturation than wild-type mice (Table 1), as previously reported12. Trf saturation decreased from 93% to 79% (Table 1) 24 h after treatment with LPS, and unlike in wild-type mice, this decrease was due to an LPS-induced increase in total iron binding capacity from 246 mg dl−1 to 263 mg dl−1. A cytokine-induced increase in serum Trf has been reported previously in mice17. We did not observe a significant change in liver nonheme iron concentrations in either group of mice after treatment with LPS. As expected12, liver nonheme iron concentrations were significantly higher in Hfe−/− mice than in wild-type mice (Table 1). Our results suggest that Hamp mediates hyposideremia in response to inflammation in an Hfe-dependent manner. The changes in iron cycling that we attribute to Hamp persist even after Hamp expression has returned to basal levels.
To test whether differential expression of other iron metabolic genes is required for the induction of hyposideremia, we treated 8-week-old female mice with LPS, killed them after 1.5 h and analyzed liver-derived cDNA by microarray. We interrogated multiple genes involved in iron metabolism, oxidative stress, hypoxia and inflammation using IronChip (version 4.0), a sensitive and accurate cDNA-based microarray platform for analyzing mouse tissue samples11. Gene symbols and accession numbers are given in the Methods section and in Table 2, and specific changes in gene expression are given in Supplementary Table 1 online.
The gene-response patterns of wild-type and Hfe-deficient mice treated with LPS were similar and consistent with innate immunity and stress responses (Fig. 3). Expression of Tnf and genes that are induced by or participate in feedback loops with Tnf increased in response to LPS, independent of genotype. Expression of hepatic Tlr2 (encoding toll-like receptor 2) mRNA increased in mice treated with LPS, consistent with a response to Tnf and Il6 (ref. 18) and signaling through the LPS receptor Tlr4 (ref. 19). Other Tnf-induced genes were Zfp36 (encoding zinc finger protein 36), Jun, Hmox1 (encoding heme oxygenase 1) and Mt1 and Mt2 (encoding metallothionein 1 and 2). Stress-induced genes included Ldh1 (encoding lactate dehydrogenase 1) and Por (encoding NADH cytochrome P450 oxidoreductase). Arntl (encoding aryl hydrocarbon receptor, nuclear translocator-like) and Clock (encoding circadian locomoter output cycles kaput), two genes that regulate circadian rhythm, were downregulated in response to LPS in wild-type and Hfe-deficient mice. We observed no substantial changes in expression of Slc39a1 (encoding solute carrier 39a1), Slc11a2 (encoding solute carrier 11a2), Heph (encoding hephaestin), Cp (encoding ceruloplasmin), Hfe, Trfr2 (encoding transferrin receptor 2), Ftl or Fth (encoding light or heavy chains of ferritin) mRNA in wild-type and Hfe-deficient mice treated with LPS (Supplementary Table 1 online). Expression of Trfr (encoding transferrin receptor) mRNA was lower in Hfe-deficient mice, regardless of LPS treatment, probably owing to regulation by the iron-responsive element in response to hepatic iron overload11. These findings suggest that Hfe-deficient mice mount a general inflammatory response to LPS without an alteration in the expression of many key iron metabolic genes.
We analyzed pools of total liver RNA from six Hfe−/− mice, six Hfe882A/882A mice or four wild-type mice challenged with LPS on IronChip (version 4.0) in comparison with pooled total liver RNA from four untreated isogenic wild-type mice. Genes that were differentially expressed between the different mice are listed. For brevity, we omitted some genes of lesser importance. The average expression changes of all differentially expressed genes with their respective standard deviations are shown in Supplementary Table 1 online.
Both Hamp and lipocalin-2 (Lcn-2) have been postulated to be involved in ACD20,21. Hamp blocks cellular iron egress5,9 and Lcn-2 may reduce free iron and participate in siderophore-mediated iron uptake22,23. Hamp expression was not substantially higher in Hfe−/− mice treated with LPS (Figs. 1 and 3), but Lcn2 expression increased in an Hfe-independent fashion (Fig. 3). This difference further distinguishes Hamp expression from the general inflammatory response.
In the past, it has been difficult to differentiate between factors that induce inflammation and factors that induce hyposideremia in response to inflammation. For example, antibodies against Tnf can ameliorate the anemia associated with rheumatoid arthritis, but they also reduce the severity of the underlying inflammatory condition24. Hfe−/− mice allow us to uncouple inflammation and hyposideremia. Hfe−/− mice mount the expected inflammatory response to LPS but do not develop hyposideremia. We attribute this difference to inappropriately low expression of Hamp. These data suggest a new role for Hfe as a mediator of innate immunity by regulating Hamp in response to inflammation. Inhibition of HFE function may be a new therapeutic approach to treating ACD, particularly because inhibition of Hfe and Hamp expression does not abrogate the full inflammatory response. Decreased HFE activity and decreased HAMP expression are unlikely to have deleterious consequences in the short term, although longstanding iron overload has been associated with an increased incidence of infection25.
Methods
Animal care.
The Hfe−/− and Hfe882A/882A mice that we used in this study have been previously described12. All mice were born and housed in the barrier facility at Children's Hospital (Boston, Massachusetts, USA). We weaned pups at ∼21 days and maintained them on Prolab RMH 3000 LabDiet (PMI Richmond), which has 380 parts per million of iron. All of our mouse procedures are in compliance with the guidelines of the Institutional Animal Care and Use Committee at Children's Hospital, Boston.
Induction of inflammation.
We gave mice one intraperitoneal injection of 5 μg of LPS (serotype O111:B4; Sigma) in pyrogen-free phosphate-buffered saline (Invitrogen). We killed the mice 1.5 h (ref. 4) or 24 h after injection and collected tissues for extraction of RNA. Alternatively, we extracted blood from the mice to collect serum for ELISA and serum iron assays and then killed them and collected liver for analysis of nonheme iron.
RNA preparation.
We collected liver tissue and immediately stored it in RNAlater (Ambion). Then we transferred the tissue from the RNAlater to 5 ml of Trizol (Invitrogen) and homogenized it three times for 20 s each using a Polytron PT2100 (Novodirect). We extracted RNA according to the manufacturer's instructions.
Northern-blot analysis.
We separated 10 μg of total RNA from individual Hfe-deficient or wild-type mice on a 1% formaldehyde agarose gel and blotted it onto a Nylon membrane (Nytran N; Schleicher and Schuell). We hybridized the membrane to radioactively labeled probes specific for Hamp and Gapd mRNA. We quantified the signals on a fluoroimager (Molecular Dynamics, now Amersham Biosciences).
Quantitative RT-PCR.
We denatured 1 μg of total RNA at 70 °C for 10 min in the presence of 70 pmol of oligo d(T) and then chilled it on ice. We synthesized cDNA using the Superscript II First Strand Synthesis System (Invitrogen) in a 20-μl reaction. We carried out an identical reaction without reverse transcriptase to verify the absence of genomic DNA. We conducted PCR on duplicate samples using the 2× SYBR Green PCR Master Mix (Applera), 1 μl of cDNA reaction, and 0.3 μM of each primer using an ABI PRISM 7000 Sequence Detection System (Applied Bioscience) with the following cycle parameters: 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 15 s at 95 °C and 1 min at 60 °C (for Il6 and Gapd) or 65 °C (for Tnf). Primer sequences are available on request.
ELISA.
We carried out ELISAs for Il6 and Tnf (R&D Systems) according to the manufacturer's instructions.
Iron assays.
We assayed serum iron concentrations using the Serum Iron/UIBC kit (ThermoDMA) according to the manufacturer's instructions and nonheme liver iron concentrations as previously described26.
cDNA microarray analysis.
The mouse IronChip (Version 4.0) contains 365 genes that encode proteins that are directly involved in iron metabolism or in interlinked pathways such as copper metabolism, nitric oxide metabolism, inflammation or the oxidative stress pathway. We verified the sequences of all genes represented on our arrays from both ends. We spotted each gene at least six times in different locations of the chip, and many genes are represented by up to five independent cDNA clones. This redundancy, together with the use of positive (spike-in) controls and negative controls in every subgrid of the array, assures high data quality27,28. Redundancy of the genes at different positions of the microarray also assures that we can accurately recognize small changes in gene expression. We described technical details of the microarray analysis elsewhere28,29. We also previously described the use of negative hybridization controls to determine the background noise of a microarray experiment and the use of spike-in controls to determine significant ratio cut-off values28,29.
Table 2 summarizes the gene symbols mentioned in the text or shown in the figures; it includes the gene name according to the International Committee on Standardized Genetic Nomenclature for Mice or the Human Genome Organization Gene Nomenclature Committee, the GenBank accession numbers and the common gene name.
Experimental strategy.
We pooled total RNA from four to six experimental mice, and analyzed it in comparison with pooled total RNA from the same tissue from four wild-type mice on the IronChip. We incorporated Cy3 fluorescent dyes into the cDNA synthesized from total RNA from wild-type mice and Cy5 fluorescent dyes into cDNA synthesized from total RNA from the experimental mice, or vice versa (dye switch). We scored only those genes that were differentially expressed in a consistent way in both experiments. We analyzed the data in Excel (Microsoft). For each individual chip, we averaged all spots representing one cDNA. For genes represented by multiple cDNA clones on the microarray platform, we calculated the average of ratios and their standard deviation. Finally, we averaged the ratios of those genes that were scored as differentially expressed in each dye switch experiment.
Statistics.
We calculated all P values using Student's t-test using Microsoft Excel.
URL.
The complete list of genes and corresponding gene accession numbers is available at http://www.embl-heidelberg.de/ExternalInfo/hentze/suppinfo.html.
Accession numbers.
GenBank: Gapd, M32599. ArrayExpress: microarray chip, features, location, associated sequences and annotation., A-MEXP-61; samples, treatments and results (raw and normalized data), E-MEXP-95.
Note: Supplementary information is available on the Nature Genetics website.
Accession codes
References
Lee, G.R. The anemia of chronic disease. Semin. Hematol. 20, 61– 80 (1983).
Krause, A. et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 480, 147– 150 (2000).
Park, C.H., Valore, E.V., Waring, A.J. & Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 276, 7806– 7810 (2001).
Pigeon, C. et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 276, 7811– 7819 (2001).
Nicolas, G. et al. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. USA 98, 8780– 8785 (2001).
Nicolas, G. et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Invest. 110, 1037– 1044 (2002).
Weinstein, D.A. et al. Inappropriate expression of hepcidin is associated with iron refractory anemia: implications for the anemia of chronic disease. Blood 100, 3776– 3781 (2002).
Nemeth, E. et al. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood 101, 2461– 2463 (2003).
Nicolas, G. et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc. Natl. Acad. Sci. USA 99, 4596– 4601 (2002).
Nicolas, G. et al. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat. Genet. 34, 97– 101 (2003).
Muckenthaler, M. et al. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat. Genet. 34, 102– 107 (2003).
Levy, J.E., Montross, L.K., Cohen, D.E., Fleming, M.D. & Andrews, N.C. The C282Y mutation causing hereditary hemochromatosis does not produce a null allele. Blood 94, 9– 11 (1999).
Bopst, M., Haas, C., Car, B. & Eugster, H.P. The combined inactivation of tumor necrosis factor and interleukin-6 prevents induction of the major acute phase proteins by endotoxin. Eur. J. Immunol. 28, 4130– 4137 (1998).
Gabay, C. & Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340, 448– 454 (1999).
Moshage, H. Cytokines and the hepatic acute phase response. J. Pathol. 181, 257– 266 (1997).
Alvarez-Hernandez, X., Liceaga, J., McKay, I.C. & Brock, J.H. Induction of hypoferremia and modulation of macrophage iron metabolism by tumor necrosis factor. Lab. Invest. 61, 319– 322 (1989).
Gordeuk, V.R., Prithviraj, P., Dolinar, T. & Brittenham, G.M. Interleukin 1 administration in mice produces hypoferremia despite neutropenia. J. Clin. Invest. 82, 1934– 1938 (1988).
Matsuguchi, T., Musikacharoen, T., Ogawa, T. & Yoshikai, Y. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J. Immunol. 165, 5767– 5772 (2000).
Fan, J. & Malik, A.B. Toll-like receptor-4 (TLR4) signaling augments chemokine-induced neutrophil migration by modulating cell surface expression of chemokine receptors. Nat. Med. 9, 315– 321 (2003).
Ganz, T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 102, 783– 788 (2003).
Kaplan, J. Mechanisms of cellular iron acquisition: another iron in the fire. Cell 111, 603– 606 (2002).
Yang, J. et al. An iron delivery pathway mediated by a lipocalin. Mol. Cell 10, 1045– 1056 (2002).
Goetz, D.H. et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033– 1043 (2002).
Davis, D. et al. Anaemia of chronic disease in rheumatoid arthritis: in vivo effects of tumour necrosis factor alpha blockade. Br. J. Rheumatol. 36, 950– 956 (1997).
Muench, K.H. Hemochromatosis and infection: alcohol and iron, oysters and sepsis. Am. J. Med. 87, 40N– 43N (1989).
Torrance, J.D. & Bothwell, T.H. Tissue iron stores. in Methods in Hematology vol. 1 (ed. Cook, J.D.) 104– 109 (Churchill Livingstone, New York, 1980).
Muckenthaler, M. et al. Relationships and distinctions in iron regulatory networks responding to interrelated signals. Blood 101, 3690– 3698 (2002).
Richter, A. et al. Comparison of fluorescent tag DNA labeling methods used for expression analysis by DNA microarrays. Biotechniques 33, 620– 628, 630 (2002).
Thompson, K. et al. Mouse brains deficient in H-ferritin have normal iron concentration but a protein profile of iron deficiency and increased evidence of oxidative stress. J. Neurosci. Res. 71, 46– 63 (2003).
Zhou, X.Y. et al. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc. Natl. Acad. Sci. USA 95, 2492– 2497 (1998).
Acknowledgements
We thank A. Donovan and F. Huang for discussions about Hfe−/− mice; B. Miñana, V. Benes, R. Carmouche and M. Benesova for maintaining the IronChip microarray platform; V. Benes for advice on quantitative RT-PCR; and E. Vainshtein for help with the submission of our microarray data to MIAMExpress. M.W.H acknowledges funds from the Gottfried Wilhelm Leibniz prize. N.C.A. is an Associate Investigator of the Howard Hughes Medical Institute and was additionally funded by a grant from the US National Institutes of Health. C.N.R. was funded by a training grant from the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Roy, C., Custodio, Á., de Graaf, J. et al. An Hfe-dependent pathway mediates hyposideremia in response to lipopolysaccharide-induced inflammation in mice. Nat Genet 36, 481–485 (2004). https://doi.org/10.1038/ng1350
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1350
This article is cited by
-
Tumour necrosis factor alpha downregulates human hemojuvelin expression via a novel response element within its promoter
Journal of Biomedical Science (2012)
-
Iron and the immune system
Journal of Neural Transmission (2011)
-
Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD
Journal of Molecular Medicine (2009)
-
Iron and inflammation: cross-talk between pathways regulating hepcidin
Journal of Molecular Medicine (2008)
-
A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells
Journal of Molecular Medicine (2008)