Abstract
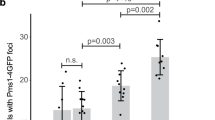
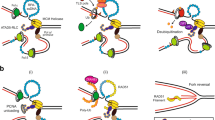
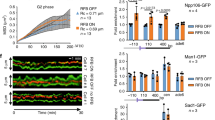
Proliferating cell nuclear antigen (PCNA) is required for mismatch repair (MMR) and has been shown to interact with complexes containing Msh2p or MLH1 (refs 1–4). PCNA has been implicated to act in MMR before and during the DNA synthesis step, although the biochemical basis for the role of PCNA early in MMR is unclear1,3,5. Here we observe an interaction between PCNA and Msh2p-Msh6p mediated by a specific PCNA-binding site present in Msh6p. An msh6 mutation that eliminated the PCNA-binding site caused a mutator phenotype and a defect in the interaction with PCNA. The association of PCNA with Msh2p-Msh6p stimulated the preferential binding of Msh2p-Msh6p to DNA containing mispaired bases. Mutant PCNA proteins encoded by MMR-defective pol30 alleles were defective for interaction with Msh2p-Msh6p and for stimulation of mispair binding by Msh2p-Msh6p. Our results suggest that PCNA functions directly in mispair recognition and that mispair recognition requires a higher-order complex containing proteins in addition to Msh2p-Msh6p.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gu, L., Hong, Y., McCulloch, S., Watanabe, H. & Li, G.M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26, 1173–1178 ( 1998).
Johnson, R.E. et al. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271, 27987–27990 (1996).
Umar, A. et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87, 65– 73 (1996).
Hughes, P., Tratner, I., Ducoux, M., Piard, K. & Baldacci, G. Isolation and identification of the third subunit of mammalian DNA polymerase delta by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res. 27, 2108–2114 (1999).
Umar, A. & Kunkel, T.A. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur. J. Biochem. 238, 297–307 ( 1996).
Warbrick, E. PCNA binding through a conserved motif. Bioessays 20 , 195–199 (1998).
Chen, C., Merrill, B.J., Lau, P.J., Holm, C. & Kolodner, R.D. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19, 7801–7815 (1999).
Amin, N.S. & Holm, C. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics 144, 479–493 (1996).
Ayyagari, R., Impellizzeri, K.J., Yoder, B.L., Gary, S.L. & Burgers, P.M. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 15 , 4420–4429 (1995).
Kokoska, R.J., Stefanovic, L., Buermeyer, A.B., Liskay, R.M. & Petes, T.D. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics 151, 511– 519 (1999).
Marsischky, G.T. & Kolodner, R.D. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274, 26668–26682 (1999).
Gradia, S., Acharya, S. & Fishel, R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91, 995 –1005 (1997).
Alani, E., Sokolsky, T., Studamire, B., Miret, J.J. & Lahue, R.S. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol. 17, 2436–2447 (1997).
Blackwell, L.J., Martik, D., Bjornson, K.P., Bjornson, E.S. & Modrich, P. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 273, 32055–32062 (1998).
Iaccarino, I., Marra, G., Palombo, F. & Jiricny, J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J. 17, 2677–2686 (1998).
Kolodner, R.D. & Marsischky, G.T. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9, 89–96 (1999).
Prolla, T.A., Pang, Q., Alani, E., Kolodner, R.D. & Liskay, R.M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265, 1091–1093 (1994).
Habraken, Y., Sung, P., Prakash, L. & Prakash, S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr. Biol. 7, 790–793 ( 1997).
Drotschmann, K., Aronshtam, A., Fritz, H.J. & Marinus, M.G. The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 26, 948– 953 (1998).
Spampinato, C. & Modrich, P. The MutL ATPase is required for mismatch repair. J. Biol. Chem. 275 , 9863–9869 (2000).
Rose, M., Winston, F. & Hieter, P. Methods in Yeast Genetics: A Laboratory Course (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1990).
Lea, D.E. & Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49, 264–268 (1949).
Flores-Rozas, H. & Kolodner, R.D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl Acad. Sci. USA 95, 12404–12409 (1998).
Marsischky, G.T., Filosi, N., Kane, M.F. & Kolodner, R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10, 407– 420 (1996).
Vallejo, A.N., Pogulis, R.J. & Pease, L.R. Mutagenesis and synthesis of novel recombination genes using PCR. in PCR Primer: A Laboratory Manual (eds Dieffenbach, C.W. & Dveksler, G.S.) 603–612 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1995).
Alani, E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 16, 5604–5615 (1996).
Fien, K. & Stillman, B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 12, 155–163 (1992).
Sia, E.A., Kokoska, R.J., Dominska, M., Greenwell, P. & Petes, T.D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17, 2851–2858 (1997)
Acknowledgements
We thank N.S. Amin, R. Das Gupta, P. Lau, T. Nakagawa and K. Schmidt for helpful discussions and comments on the manuscript, and J. Weger and J. Green for DNA sequencing. This work was supported by National Institutes of Health grant GM50006 to R.D.K. and a fellowship from the Jane Coffin Childs Memorial Fund to H.F.-R.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Flores-Rozas, H., Clark, D. & Kolodner, R. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet 26, 375–378 (2000). https://doi.org/10.1038/81708
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/81708
This article is cited by
-
Mispair-bound human MutS–MutL complex triggers DNA incisions and activates mismatch repair
Cell Research (2021)
-
Interface of DNA Repair and Metabolism
Current Tissue Microenvironment Reports (2020)
-
Identification of a novel large EPCAM-MSH2 duplication, concurrently with LOHs in chromosome 20 and X, in a family with Lynch syndrome
International Journal of Colorectal Disease (2019)
-
MutSβ promotes trinucleotide repeat expansion by recruiting DNA polymerase β to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis
Cell Research (2016)
-
Epigenetic Enhancement of the Post-replicative DNA Mismatch Repair of Mammalian Genomes by a Hemi-mCpG-Np95-Dnmt1 Axis
Scientific Reports (2016)