Abstract
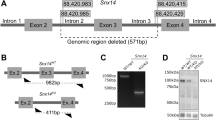
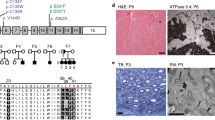
Tay–Sachs and Sandhoff diseases are clinically similar neurodegenerative disorders. These two sphingolipidoses are characterized by a heritable absence of β–hexosaminidase A resulting in defective GM2 ganglioside degradation. Through disruption of the Hexa and Hexb genes in embryonic stem cells, we have established mouse models corresponding to each disease. Unlike the two human disorders, the two mouse models show very different neurologic phenotypes. Although exhibiting biochemical and pathologic features of the disease, the Tay–Sachs model showed no neurological abnormalities. In contrast, the Sandhoff model was severely affected. The phenotypic difference between the two mouse models is the result of differences in the ganglioside degradation pathway between mice and humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Gravel, R.A. et al. The GM2 gangliosidoses. in The metabolic and molecular bases of inherited disease (eds. Scriver, C.R., A.L. Beaudet, W. S. Sly, & D. Valle) 2839–2879 (McGraw-Hill Inc., 1995).
Leinekugel, P., Michel, S., Conzelmann, E. & Sandhoff, K. Quantitative correlation between the residual activity of β-hexosaminidase A and arylsulfatase A and the severity of the resulting lysosomal storage disease. Hum. Genet. 88, 513–523 (1992).
Yamanaka, S. et al. Targeted disruption of the Hexa gene results in mice with biochemical and pathologic features of Tay-Sachs disease. Proc. natn. Acad. Sci. U.S.A. 91, 9975–9979 (1994).
Taniike, M. et al. Neuropathology of mice with targeted disruption of Hexa gene, a model of Tay-Sachs disease. Acta Neuropathol. 89, 296–304 (1995).
Yamanaka, S., Johnson, O.J., Norflus, F., Boles, D.J. & Praia, R.L. Structure and expression of the mouse (β-hexosaminidase genes, Hexa and Hexb. Genomics 21, 588–596 (1994).
Capecchi, M.R. Altering the genome by homologous recombination. Science 244, 1288–1292 (1989).
Lee, K.-F. et al. Targeted mutation of the gene encoding the low affinity receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69, 737–749 (1992).
Sandhoff, K., Harzer, K., Wässle, W. & Jatzkewitz, H. Enzyme alterations and lipid storage in three variants of Tay-Sachs disease. J. Neurochem. 18, 2469–2489 (1971).
Sandhoff, K. The biochemistry of sphingolipid storage diseases. Angew. Chem. Int. Ed. Engl. 16, 273–285 (1977).
Sonderfeld, S. et al. Incorporation and metabolism of ganglioside GM2 in skin fibroblasts from normal and GM2 gangliosidosis subjects. Eur. J. Biochem. 149, 247–255 (1985).
Conzelmann, E. & Sandhoff, K. Purification and characterization of an activator protein for the degradation of glycolipids GM2 and GA2 by hexosaminidase A. Hoppe-Seyler's Z. physiol. Chem. 360, 1837–1849 (1979).
Suzuki, K. Neuropathology of late onset gangliosidoses. Dev. Neurosci. 13, 205–210 (1991).
Burg, J., Banerjee, A., Conzelmann, E. & Sandhoff, K. Activating proteins for ganglioside GM2 degradation by β-hexosaminidase isoenzymes in tissue extracts from different species. Hoppe-Seyler's Z. physiol. Chem. 364, 821–829 (1983).
Sandhoff, K. & Jatzkewitz, H. A particle-bound sialyl lactosidoceramide splitting mammalian sialidase. Biochim. Biophys. Acta 141, 442–444 (1967).
Riboni, L., Caminiti, A., Bassi, R. & Tettamanti, G. The degradative pathway of gangliosides GM1 and GM2 in Neuro2a cells by sialidase. J. Neurochem. 64, 451–454 (1995).
Suzuki, Y., Jacob, J.C., Suzuki, K., Kutty, K.M. & Suzuki, K. GM2-gangliosidosis with total hexosaminidase deficiency. Neurol. 21, 313–328 (1971).
Conzelmann, E. & Sandhoff, K. Biochemical basis of late-onset neurolipidoses. Dev. Neurosci. 13, 197–204 (1991).
van den Eijnden, D.H. Chromatographic separation of gangliosides on precoated silicagel thin-layer plates. Hoppe-Seyler's Z. physiol. Chem. 352, 1601–1602 (1971).
Robertson, E.J. Embryo-derived stem cells, in Teratocarcinomas and embryonic stem cells. (ed Robertson, E. J.) (IRL Press, Oxford, 1987).
Schwarzmann, G. A simple and novel method for tritium labeling of gangliosides and other sphingolipids. Biochim. Biophys. Acta 529, 106–114 (1978).
Klima, H. et al. Over-expression of a functionally active human GM2-activator protein in Escherichia coli. Biochem. J. 292, 571–576 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sango, K., Yamanaka, S., Hoffmann, A. et al. Mouse models of Tay–Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat Genet 11, 170–176 (1995). https://doi.org/10.1038/ng1095-170
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng1095-170
This article is cited by
-
Novel diagnostic and therapeutic techniques reveal changed metabolic profiles in recurrent focal segmental glomerulosclerosis
Scientific Reports (2021)
-
Glycosphingolipids and neuroinflammation in Parkinson’s disease
Molecular Neurodegeneration (2020)
-
GM2 ganglioside accumulation causes neuroinflammation and behavioral alterations in a mouse model of early onset Tay-Sachs disease
Journal of Neuroinflammation (2020)
-
Upregulating β-hexosaminidase activity in rodents prevents α-synuclein lipid associations and protects dopaminergic neurons from α-synuclein-mediated neurotoxicity
Acta Neuropathologica Communications (2020)
-
A novel gene editing system to treat both Tay–Sachs and Sandhoff diseases
Gene Therapy (2020)