Abstract
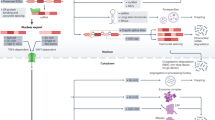
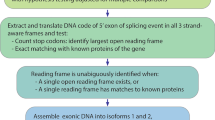
Although nonsense mutations have been associated with the skipping of specific constitutively spliced exons in selected genes, notably the fibrillin gene, the basis for this association is unclear. Now, using chimaeric constructs in a model in vivo expression system, premature termination codons are identified as determinants of splice site selection. Nonsense codon recognition prior to RNA splicing necessitates the ability to read the frame of precursor mRNA in the nucleus. We propose that maintenance of an open reading frame can serve as an additional level of scrutiny during exon definition. This process may have pathogenic and evolutionary significance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Krawczak, M., Reiss, J. & Cooper, D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90, 41–54 (1992).
Mount, S.M. A catalogue of splice junction sequences. Nucl. Acids Res. 10, 459–472 (1982).
Oshima, Y. & Qotoh, Y.J. Signals for the selection of a splice site in pre-mRNA. J. molec. Biol. 196, 247–259 (1987).
Sharp, P.A. Splicing of messenger RNA precursors. Science 235, 766–771 (1987).
Shapiro, M.B. & Senapathy, P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucl. Acids Res. 15, 7155–7175 (1987).
Grabowski, P.J., Seller, S.R. & Sharp, P.A. A multi-component complex is involved in the splicing of messenger RNA precursors. Cell 42, 345 (1985).
Padgett, R.A., Grabowski, P.J., Konarska, M.M., Seller, S.R. & Sharp, P.A. Splicing of messenger RNA precursors. A. Rev. Biochem. 55, 1119–1150 (1986).
Maniatis, T. & Reed, R. The role of small nuclear ribo nucleo proteln particles in pre-mRNA splicing. Nature 325, 673–678 (1987).
Robberson, B.L., Cote, Q.J. & Berget, S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Molec. Cell. Biol. 10, 84–94 (1990).
Niwa, M., MacDonald, C.C. & Berget, S.M. Are vertebrate exons scanned during splice-site selection? Nature 360, 277–280 (1992).
Reed, R. & Maniatis, T. A role for exon sequences and splice-site proximity in splice-site selection. Cell 46, 681–690 (1986).
Libri, D., Qoux-Pelletan, M., Brody, E. & Fiszman, M.V. Exon as well as intron sequences are cis-regulating elements for mutually exclusive alternative splicing of the β-tropomyosin gene. Molec Cell. Biol. 10, 5036–5046 (1990).
Domenjoud, L., Gallinaro, H., Kister, L., Meyer, S. & Jacob, M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and cryptic 5′ splice site. Molec. Cell. Biol. 11, 4581–4590 (1991).
Cooper, T.A. In vitro splicing of cardiac troponin T precursors. J. Biol. Chem. 267, 5330–5338 (1992).
Steingrimsdottir, H., Rowley, G., Dorado, G., Cole, J. & Lehmann, A.R. Mutations which alter splicing in the human hypoxanthine guanine phosphoribosyltransferase gene. Nucl. Acids Res. 20, 1201–1208 (1992).
Dietz, H.C. et al. The skipping of constitutive exons in vivo induced by nonsense mutations. Science 250, 680–683 (1993).
Naeger, L.K., Schoberg, R.V., Zhao, Q., Tullis, G.E. & Pintell, D.J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6, 1107–1119 (1992).
Naylor, J.A., Green, P.M., Rizza, C.R. & Gianelli, F. Analysis of factor VIII mRNA reveals defects in everyone of 28 haemophilia A patients. Hum. Molec. Genet. 2, 11–17 (1993).
Gibson, R.A., Hajianpour, A., Murer-Orlando, M., Buchwald, M. & Mathew, C.G. A nonsense mutation and exon skipping in the Fanconi anaemia group C gene. Hum. Molec. Genet. 2, 797–799 (1993).
Bach, G., Moskowitz, S.M., Phuong, T.T., Matynia, A. & Neufeld, E.F. Molecular analysis of Hurler syndrome in Druze and Muslim Arab patients in Israel: multiple allelic mutations of the IDUA gene in a small geographic area. Am. J. Hum. Genet. 53, 330–338 (1993).
Pereira, L. et al. Genomic organization of the sequence coding for fibrillin, thedefective gene product in Marfan syndrome. Hum. Molec Genet. 2, 961–968 (1993).
Dietz, H.C. et al. Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics 17, 468–475 (1993).
Mitchell, G.A. et al. Human ornithine δ-aminotransferase: cDNA cloning and analysis of the structural gene. J. Biol. Chem. 263, 14288–14295 (1988).
Black, D.L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-nueronal cells? Genes Dev. 5, 389–402 (1991).
Daar, I.O. & Maquat, L.E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Molec. Cell. Biol. 8, 802–813 (1988).
Urlaub, G., Mitchell, P.J., Ciudad, C.J. & LA Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Molec. Cell. Biol. 9, 2868–2880 (1989).
Cheng, J., Fogel-Petrovic, M. & Maquat, L.E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Molec. Cell. Biol. 10, 5215–5225 (1990).
Baserga, S.J. & Benz, E.J. Jr. B-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc. Natl. Acad. Sci. U.S.A. 89, 2935–2939 (1992).
Cheng, J. & Maquat, L.E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Molec. Cell. Biol. 13, 1892–1902 (1993).
Belgrader, P., Cheng, J. & Maquat, L.E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. U.S.A. 90, 482–486 (1993).
Carothers, A.M., Urlaub, G., Greenberger, D. & Chasin, L.A. Splicing mutants and their second-life suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Molec. Cell. Biol. 13, 5085–5098 (1993).
Kessler, O., Jiang, Y. & Chasin, L.A. Order of intron removal during splicing of endogenous adenine phosphoribosyltranferase and dihydrofolate reductase pre-mRNA. Molec. Cell. Biol. 13, 6211–6222 (1993).
Fisher, C.W. et al. Occurrence of a 2-bp (AT) deletion allele and a nonsense (G-to-T) mutant allele at E2 (DBT) locus of six patients with Maple Syrup Urine Disease: Multiple-exon skipping as a secondary effect of the mutations. Am. J. Hum. Genet. 52, 414–424 (1993).
Hawkins, J.D. A survey on intron and exon lengths. Nucl. Acids Res. 16, 9893–9908 (1988).
Dietz, H.C. et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 352, 337–339 (1991).
Dietz, H.C. et al. Marfan phenotype variability in a family segregating a missense mutation in the epidermal growth factor-like motif of the fibrillin gene. J. clin. Invest. 89, 1674–1680 (1992).
Dietz, H.C. et al. Clustering of fibrillin (FBN1) missense mutations in Marfan syndrome patients at cysteine residues in EGF-like domains. Hum. Mutat. 1, 366–374 (1992).
Hamosh, A. et al. Severe deficiency of cystic fibrosis transmembrane conductance regulator messenger RNA carrying nonsense mutation R553X and W1316X in respiratory epithelial cells of patients with cystic fibrosis. J. clin. Invest. 88, 1880–1885 (1991).
Deng, W.P. & Nickoloff, J.A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Analyt. Biochem. 200, 81–88 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Dietz, H., Kendzior, R. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet 8, 183–188 (1994). https://doi.org/10.1038/ng1094-183
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng1094-183
This article is cited by
-
RNA splicing promotes translation and RNA surveillance
Nature Structural & Molecular Biology (2005)
-
Nonsense-Associated Alternative Splicing of the Human Thyroglobulin Gene
Molecular Diagnosis (2005)
-
Messenger-RNA-binding proteins and the messages they carry
Nature Reviews Molecular Cell Biology (2002)
-
Listening to silence and understanding nonsense: exonic mutations that affect splicing
Nature Reviews Genetics (2002)
-
The power of point mutations
Nature Genetics (2001)