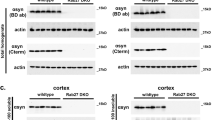
Abstract
The gracile axonal dystrophy (gad) mouse is an autosomal recessive mutant that shows sensory ataxia at an early stage, followed by motor ataxia at a later stage1. Pathologically, the mutant is characterized by 'dying-back' type axonal degeneration and formation of spheroid bodies in nerve terminals2,3,4,5. Recent pathological observations have associated brain ageing and neurodegenerative diseases with progressive accumulation of ubiquitinated protein conjugates6,7. In gad mice, accumulation of amyloid β-protein and ubiquitin-positive deposits occur retrogradely along the sensory and motor nervous systems8,9. We previously reported that the gad mutation was transmitted by a gene on chromosome 5 (refs 10,11). Here we find that the gad mutation is caused by an in-frame deletion including exons 7 and 8 of Uchl1, encoding the ubiquitin carboxy-terminal hydrolase (UCH) isozyme (Uch-l1) selectively expressed in the nervous system and testis12,13,14,15. The gad allele encodes a truncated Uch-l1 lacking a segment of 42 amino acids containing a catalytic residue16. As Uch-l1 is thought to stimulate protein degradation by generating free monomeric ubiquitin16,17,18, the gad mutation appears to affect protein turnover. Our data suggest that altered function of the ubiquitin system directly causes neurodegeneration. The gad mouse provides a useful model for investigating human neurodegenerative disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yamazaki, K. et al. Gracile axonal dystrophy (GAD), a new neurological mutant in the mouse. Proc. Soc. Exp. Biol. Med. 187, 209–215 (1988).
Mukoyama, M., Yamazaki, K., Kikuchi, T. & Tomita, T. Neuropathology of gracile axonal dystrophy (GAD) mouse (an animal model of central axonopathy in primary sensory neurons). Acta Neuropathol. 79, 294–299 ( 1989).
Kikuchi, T., Mukoyama, M., Yamazaki, K. & Moriya, H. Axonal degeneration of ascending sensory neurons in gracile axonal dystrophy mutant mouse. Acta Neuropathol. (Berl.) 80, 145–151 (1990).
Oda, K., Yamazaki, K., Miura, H., Shibasaki, H. & Kikuchi, T. Dying back type axonal degeneration of sensory nerve terminals in muscle spindles of the gracile axonal dystrophy (GAD) mutant mouse. Neuropathol. Appl. Neurobiol. 18, 265–281 (1992).
Miura, H. et al. Progressive degeneration of motor nerve terminals in GAD mutant mouse with hereditary sensory axonopathy. Neuropathol. Appl. Neurobiol. 19, 41–51 ( 1993).
Arnold, J. et al. Ubiquitin and its role in neurodegeneration. Prog. Brain Res. 117, 23–34 (1998).
Alves-Rodrigues, A., Gregori, L. & Figueiredo-Pereira, M.E. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 21, 516–520 (1998).
Ichihara, N. et al. Axonal degeneration promotes abnormal accumulation of amyloid β-protein in ascending gracile tract of gracile axonal dystrophy (GAD) mouse. Brain Res. 695, 173–178 (1995).
Wu, J. et al. Abnormal ubiquitination of dystrophic axons in central nervous system of gracile axonal dystrophy (GAD) mutant mouse. Alzheimer's Res. 2, 163–168 ( 1996).
Yamazaki, K., Sakakibara, A., Tomita, T., Mukoyama, M. & Kikuchi, T. Location of gracile axonal dystrophy (gad) on chromosome 5 of the mouse. Jpn. J. Genet. 62, 479–484 (1987).
Suh, J.G., Yamanishi, T., Matsui, K., Tanaka, K. & Wada, K. Mapping of the gracile axonal dystrophy (gad) gene to a region between D5Mit197 and D5Mit113 on proximal mouse chromosome 5. Genomics 27, 549– 551 (1995).
Day, I.N.M. & Thompson, R.J. Molecular cloning of cDNA coding for human PGP 9.5 protein. A novel cytoplasmic marker for neurones and neuroendocrine cells. FEBS Lett. 210, 157– 160 (1987).
Wilkinson, K.D. et al. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246, 670– 673 (1989).
Day, I.N.M., Hinks, L.J. & Thompson, R.J. The structure of the human gene encoding protein gene product 9.5 (PGP9.5), a neuron-specific ubiquitin C-terminal hydrolase. Biochem. J. 268, 521–524 (1990).
Kajimoto, Y. et al. cDNA cloning and tissue distribution of a rat ubiquitin carboxyl-terminal hydrolase PGP9.5. J. Biochem. 112, 28– 32 (1992).
Larsen, C.N., Price, J.S. & Wilkinson, K.D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry 35, 6735–6744 ( 1996).
Dang, L.C., Melandri, F.D. & Stein, R.S. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amino-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868– 1879 (1998).
Larsen, C.N., Krantz, B.A. & Wilkinson, K.D. Substrate specificity of deubiquitinating enzyme: ubiquitin C-terminal hydrolases. Biochemistry 37, 3358–3368 (1998).
Dietrich, W.F. et al. A genetic map of the mouse with 4,006 simple sequence length polymorphism. Nature Genet. 7, 220– 245 (1994).
Wakana, S., Shiroishi, T., Puschel, A.W. & Imai, K. The mouse semaphorin F (Semaf) maps to chromosome 15. Mamm. Genome 8, 698–699 ( 1997).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Mayer, A.N. & Wilkinson, K.D. Detection, resolution, and nomenclature of multiple ubiquitin carboxyl-terminal esterases from bovine calf thymus. Biochemistry 28, 166–172 (1989).
Wilkinson, K.D., Deshpande, S. & Larsen, C.N. Comparisons of neuronal (PGP 9.5) and non-neuronal ubiquitin C-terminal hydrolases. Biochem. Soc. Trans. 20, 631–637 (1992).
Ii, K., Ito, H., Tanaka, K. & Hirano, A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J. Neuropathol. Exp. Neurol. 56, 125–131 (1997).
Leroy, E. et al. The ubiquitin pathway in Parkinson's disease. Nature 395, 451–452 ( 1998).
Ophoff, R.A., Terwindt, G.M., Frants, R.R. & Ferrari, M.D. P/Q-type Ca2+ channel defects in migraine, ataxia and epilepsy. Trends Pharmacol. Sci. 19, 121– 127 (1998).
Lowe, J., McDermott, H., Landon, M., Mayer, R.J. & Wilkinson, K.D. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J. Pathol. 161, 153–160 ( 1990).
Bizzi, A., Schaetzle, B., Patton, A., Gambetti, P. & Autilio-Gambetti, L. Axonal transport of two major components of the ubiquitin system: free ubiquitin and ubiquitin carboxyl-terminal hydrolase PGP 9.5. Brain Res. 548 292– 299 (1991).
Wenthold, R.J., Yokotani, N., Doi, K. & Wada, K. Immunochemical characterization of the non-NMDA glutamate receptor using subunit-specific antibodies. J. Biol. Chem. 267, 501–507 (1992).
Harada, T., Harada, C., Sekiguchi, M. & Wada, K. Light-induced retinal degeneration suppresses developmental progression of flip-to-flop alternative splicing in GluR1. J. Neurosci. 18, 3336–3343 (1998).
Acknowledgements
We thank Y. Hayashizaki, T. Tomita, K. Oda and K. Tanaka for helpful advice; R.S. Petralia for critical reading of our manuscript; T. Shiroishi for supplying M. m. molossinus; and M. Shikama for the breeding and care of animals. This work was supported in part by research grants from the Ministry of Education, Science, Sports and Culture, the Ministry of Health and Welfare, and the Science and Technology Agency of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saigoh, K., Wang, YL., Suh, JG. et al. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet 23, 47–51 (1999). https://doi.org/10.1038/12647
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/12647
This article is cited by
-
Animal models of Parkinson’s disease: bridging the gap between disease hallmarks and research questions
Translational Neurodegeneration (2023)
-
Podocyte-specific deletion of ubiquitin carboxyl-terminal hydrolase L1 causes podocyte injury by inducing endoplasmic reticulum stress
Cellular and Molecular Life Sciences (2023)
-
Deubiquitinating enzymes (DUBs): decipher underlying basis of neurodegenerative diseases
Molecular Psychiatry (2022)
-
Structural basis for specific inhibition of the deubiquitinase UCHL1
Nature Communications (2022)
-
Neonatal 6-hydroxydopamine lesioning of rats and dopaminergic neurotoxicity: proposed animal model of Parkinson’s disease
Journal of Neural Transmission (2022)