Abstract
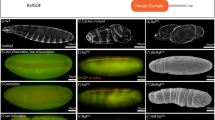
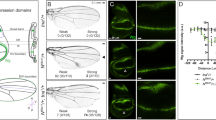
In both vertebrate and invertebrate development, cells are often programmed to adopt fates distinct from their neighbors. Genetic analyses in Drosophila melanogaster have highlighted the importance of cell surface and secreted proteins in these cell fate decisions. Homologues of these proteins have been identified and shown to play similar roles in vertebrate development1–5. Fringe, a novel signalling protein, has been shown to induce wing margin formation in Drosophila6. Fringe shares significant sequence homology and predicted secondary structure similarity with bacterial glycosyltransferases7. Thus, fringe may control wing development by altering glycosylation of cell surface and/or secreted molecules. Recently, two fringe genes were isolated from Xenopus laevis8. We report here the cloning and characterization of three murine fringe genes (lunatic fringe, manic fringe and radical fringe). We find in several tissues that fringe expression boundaries coincide with Notch-dependent patterning centres and with Notch-ligand expression boundaries. Ectopic expression of murine manic fringe or radical fringe in Drosophila results in phenotypes that resemble those seen in Notch mutants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Parr, B.A. & McMahon, A.P. Wnt genes and vertebrate development. Curr. Opin. Genet Dev. 4, 523–528 (1994).
Ingham, P.W. Signaling by hedgehog-family proteins in Drosophila and vertebrate development. Curr. Opin. Genet. Dev. 5, 492–498 (1995).
Hogan, B.L.M. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10, 1580–1394 (1996).
Lardelli, M., Williams, R. & Lendahl, U. Notch-related genes in animal development. Int. J. Dev. Biol. 39, 769–780 (1995).
Kopan, R. & Turner, D.L., Notch pathway: democracy and aristocracy in the selection of cell fate. Curr. Opin. Neurobiol. 6, 594–601 (1996).
Irvine, K.D. & Wieschaus, E., Fringe, a boundary-specific molecule, mediates interactions between dorsal and ventral cells during. Drosophila wing development. Cell 79, 595–606 (1994).
Yuan, Y.P., Schultz, J., Mlodzik, M. & Bork, P., Fringe-like signaling molecules may be glycosyItransf erases. Cell 88, 9–11 (1997).
Wu, J.Y., Zhang, W.-J. & Rao, Y. The s jcreted product of Xenopus gene lunatic Fringe, a vertebrate signaling molecule. .Science 273, 355–358 (1996).
Conlon, R.A., Reaume, A.G. & Rossant, J. Notchl is required for the coordinate segmentation of somites. Development 121, 1533–1545 (1995).
Bettenhausen, B., Hrabe de Angelis, M., Simon, D., Guenet, J.-L. & Gossler, A. Transient and restricted expression during mou ;e embryogenesis of DII1, a murine gene closely related to Drosophila Delta. Development 121, 2407–2418 (1995).
Lindsell, C.E., Shawber, C.J., Boulter, J. & Weinmaster, G. Jagged: a mammalian ligand that activates Notch1. Cell 80, 909–917 (1995).
Myat, A., Henrique, D., Ish-Horowicz, D. & Lewis, J. A chick homologue of Serrate and its relationship with Notch and De ta homologues during central neurogenesis. Dev. Biol. 174, 233–247 (1996).
Shawber, C., Boulter, J., Lindsell, C.E. & Weinmaster, G. Jagged2: A serrate-like gene expressed during rat embryogenesis. Dev. Biol. 180, 370–376 (1996).
Kim, J., Irvine, K.D. & Carrol, S.B. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell 82, 795–802 (1995).
Brand, A.M. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Hinz, U., Giebel, B. & Campos-Ortega, J.A. Tho basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell 76, 77–87 (1994).
Gustafson, K. & Boulianne, G.L. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39, 174–182 (1996).
Yen, E., Gustafson, K. & Boullianne, G.L. Green f uorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 92, 7036–7040 (1995).
Fleming, R.J., Scottgale, T.N., Diederich, R.J. & Artavanis-Tsakonas, S. The gene Serrate encodes a putative EGF-like transmem brane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 4, 2188–2201 (1990).
Thomas, U., Speicher, S.A. & Knust, E., The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development 11, 749–761 (1991).
Speicher, S.A., Thomas, U., Hinz, U. & Knust, E., Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development 120, 535–544 (1994).
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M.E. Notch signaling. Science 268, 225–232 (1995).
Vassin, H., Bremer, K.A., Knust, E. & Campos-Ortega, J.A. The neurogenic genes Delta of Drosophila melanogaster 63, 431–440 (1987).
Kopczynski, C.C., Alton, A.K., Fechtel, K., & Muskavitch, M.A.T. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 2, 1723–1735 (1988).
Parks, A.L. & Muskavitch, M.A.T. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev. Biol. 157, 484–496 (1993).
Parody, T.R. & Muskavitch, M.A.T. The pleiotropic function of Delta during postembryonic development of Drosophila melanogaster. Genetics 135, 527–539 (1993).
Muskavitch, M.A.T. Delta-Notch signaling and Drosophila cell fate choice. Dev. Biol. 166, 415–430 (1994).
Robey, E. et al. An activated form of Notch influences the choice between CD4 and CDS T cell lineages. Cell 87, 483–492 (1996).
Rodriguez-Esteban, C. et al. Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature 386, 360–366 (1997).
Laufer, E. et al. Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature 386, 366–373 (1997).
de Angelis, M.H., Mclntyre II, J. & Gossler, A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386, 717–721 (1997).
Johnston, S.H. et al. A family of mammalian fringe genes implicated in boundary determination and the Notch pathway. Development 124, 2245–2254 (1997).
Hui, C.-C., Slusarski, D., Platt, K.A., Holmgren, R & Joyner, A.L. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 162, 402–413 (1994).
Spradling, A.C. P-element-mediated transformation in Drosophila: A practical approach (ed. Roberts, D.B.) 175–197 (IRL Press, Oxford, 1986).
Lawrence, P.A., Johnston, P. & Morata, G. Methods of marking cells, in Drosophila: A Practical Approach, (ed. Roberts, D.B.) 229–242 (IRL Press, Oxford, 1986).
Tomlinson, A. & Ready, D.F. Cell fate in the Drosophila ommatidium. Dev. Biol. 123, 264–275 (1987).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cohen, B., Bashirullah, A., Dagnino, L. et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet 16, 283–288 (1997). https://doi.org/10.1038/ng0797-283
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0797-283
This article is cited by
-
Glycosylation in cancer: mechanisms and clinical implications
Nature Reviews Cancer (2015)
-
Myeloproliferation and hematopoietic stem cell dysfunction due to defective Notch receptor modification by O-fucose glycans
Seminars in Immunopathology (2012)
-
Compartmentalised expression of Delta-like 1 in epithelial somites is required for the formation of intervertebral joints
BMC Developmental Biology (2007)
-
Notch1 expression and ligand interactions in progenitor cells of the mouse olfactory epithelium
Journal of Molecular Histology (2007)
-
Upstream Regulatory Region of Zebrafish lunatic fringe: Isolation and Promoter Analysis
Marine Biotechnology (2006)