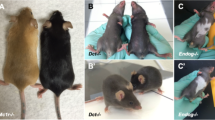
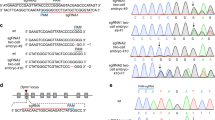
Abstract
The beige mutation is a murine autosomal recessive disorder, resulting in hypopigmentation, bleeding and immune cell dysfunction. The gene defective in beige is thought to be a homologue of the gene for the human disorder Chediak–Higashi syndrome. We have identified the murine beige gene by in vitro complementation and positional cloning, and confirmed its identification by defining mutations in two independent mutant alleles. The sequence of the beige gene message shows strong nucleotide homology to multiple human ESTs, one or more of which may be associated with the Chediak–Higashi syndrome gene. The amino acid sequence of the Beige protein revealed a novel protein with significant amino acid homology to orphan proteins identified in Saccharomyces cerevisiae, Caenorhabditis elegans and humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Spicer, S.S., Sato, A., Vincent, R., Eguchi, M. & Poon, K.C. Lysosome enlargement in the Chediak-Higashi syndrome. Fed. Proc. 40, 1451–1455 (1981).
Lutzner, M.A., Tierney, J.H. & Benditt, E.R. Giant granules and widespread cytoplasmic inclusions in a genetic syndrome of Aleutian Mink. Lab. Invest. 14, 2063–2079 (1966).
Kramer, J.W., Davis, W.C. & Prieur, D.J., The Chediak-Higashi syndrome of cats. Lab. Invest. 36, 554 (1977).
Padget, G.A., Leader, R.W., Gorham, J.R. & O'Mary, C.C. The familial occurrence of the Chediak-Higashi syndrome in mink and cattle. Genetics 49, 505–512 (1964).
Taylor, R.F. & Farrell, R.K. Light and electron microscopy of peripheral blood neutrophils in a killer whale affected with Chediak-Higashi syndrome. Fed. Proc. Fed. Am. Soc. Exp. Biol. 32, 822A (1973).
Perou, C.M. & Kaplan, J. Complementation analysis of Chediak-Higashi Syndrome: the same gene may be responsible for the defect in all patients and species. Somat. Cell Mol. Genet. 19, 459–468 (1993).
Perou, C.M., Justice, M.J., Pryor, R.J. & Kaplan, J. Complementation of the Beige mutation in cultured cells by episomally-replicating murine yeast artificial chromosomes. Proc. Natl. Acad. Sci. USA 93, 5905–5909 (1996).
Novak, E.K., Hui, S.-W. & Swank, R.T. Platelet storage pool deficiency in mouse pigment mutations associated with seven distinct genetic loci. Blood 63, 536–544 (1984).
White, R.A. et al. The murine pallid mutation is a platelet storage pool disease associated with the protein 4.2 (pallidin) gene. Nature Genet. 2, 80–83 (1992).
Haak, R.A., Ingraham, L.A., Baehner, R.L. & Boxer, L.A. Membrane fluidity in humans and mouse Chediak-Higashi leukocytes. J. Clin. Invest. 64, 138–143 (1979).
Oliver, J.M., Zurier, R.B. & Berlin, R.D. Concanavalin A cap formation on polymorphonuclear leukocytes of normal and beige (Chediak-Higashi) mice. Nature 253, 471–473 (1975).
Perou, C.M. & Kaplan, J. Chediak-Higashi Syndrome is not due to a defect in microtubule-based lysosomal mobility. J. Cell Sci. 106, 99–107 (1993).
Oliver, J.M., Krawiec, J.A. & Berlin, R.D. Carbamycholine prevents giant granule formation in cultured fibroblasts from beige (Chediak-Higashi) mice. J. Cell Biol. 69, 205–210 (1976).
Gish, W. & States, D.J. Identification of protein coding regions by database similarity search. Nature Genet. 3, 266–272 (1993).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Rodriguez, F., Martegani, E., Mauri, I. & Alberghina, L. The sequence of 8.8kb of yeast chromosome III cloned in lambda PM3270 contains an unusual long ORF (YCR601). Yeast 7, 631–641 (1991).
Wilson, R. et al. 2.2Mb of contiguous nucleotide sequence from chromosome III of C.elegans . Nature 368, 32–38 (1994).
Feuchter, A.E., Freeman, J.D. & Mager, D.L. Strategy for detecting cellular transcripts promoted by human endogenous long terminal repeats: identification of a novel gene (CDC4L) with homology to yeast CDC4. Genomics 13, 1237–1246 (1992).
van der Voorn, L. & Ploegh, H.L. The WD-40 Repeat. FEBS Lett. 307, 131–134 (1992).
Duronio, R.J., Gordon, J.I. & Boguski, M.S. Comparative analysis of the beta-transducin family with identification of several new members including PWP1, a nonessential gene of Saccharomyces cerevisiae that is diveigently transcribed from NMT1. Proteins 13, 41–66 (1992).
Wang, D.S., Shaw, R., Winkelmann, J.C. & Shaw, G. Binding of PH domains of beta-adrenergic receptor kinase and beta-spectrin to WD40/beta-transducin repeat containing regions of the beta-subunit of trimeric G-proteins. Biochem. Biophys. Res. Comm. 203, 29–35 (1994).
Kelley, E.M. Mouse News Lett. 16, 36 (1957).
Bucci, C. et al. The small GTPase rab5 functions as a regulatory factor in the eariy endocytic pathway. Cell 70, 715–728 (1992).
Zerial, M. & Stenmark, H. GTPases in vesicular transport. Curr. Opin. Cell Biol. 5, 613–620 (1993).
Colombo, M.I., Mayorga, L.S., Nishimoto, I., Ross, E.M. & Stahl, P.D. Gs regulation of endosome fusion suggests a role for signal transduction pathways in endocytosis. J. Biol. Chem. 269, 14919–14923 (1994).
Balch, W.E., GTP-binding proteins in vesicular transport. TIBS 15, 473–477 (1990).
Elazar, Z. et al. ADP-ribosylatton factor and coatamer couple fusion to vesicle budding. J. Cell Biol. 124, 415–424 (1994).
Gnirke, A., Huxley, C., Peterson, K. & Olson, M. Microinjection of Intact 200- to 500-kb Fragments of YAC DNA into Mammalian Cells. Genomics 9, 742–750 (1991).
Jenkins, N.A., Copeland, N.G., Taylor, B.A. & Lee, B.K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus . J. Virol. 43, 26–36 (1982).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Perou, C., Moore, K., Nagle, D. et al. Identification of the murine beige gene by YAC complementation and positional cloning. Nat Genet 13, 303–308 (1996). https://doi.org/10.1038/ng0796-303
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0796-303
This article is cited by
-
Dendritic cells in cancer immunology
Cellular & Molecular Immunology (2022)
-
Mouse models and strain-dependency of Chédiak-Higashi syndrome-associated neurologic dysfunction
Scientific Reports (2019)
-
Genome-wide screening of mouse knockouts reveals novel genes required for normal integumentary and oculocutaneous structure and function
Scientific Reports (2019)
-
Towards the targeted management of Chediak-Higashi syndrome
Orphanet Journal of Rare Diseases (2014)
-
MULT1E/mIL-12: a novel bifunctional protein for natural killer cell activation
Gene Therapy (2014)