Abstract
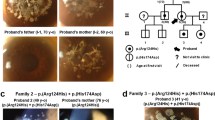
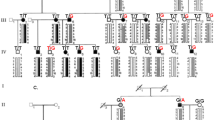
The intermediate filament cytoskeleton of corneal epithelial cells is composed of cornea-specific keratins K3 and K12 (refs 1,2). Meesmann's corneal dystrophy (MCD) is an autosomal dominant disorder causing fragility of the anterior corneal epithelium3,4, where K3 and K12 are specifically expressed5. We postulated that dominant-negative mutations in these keratins might be the cause of MCD. K3 was mapped to the type-ll keratin gene cluster on 12q; and K12 to the type-l keratin cluster on 17q using radiation hybrids. We obtained linkage to the K12 locus in Meesmann's original German kindred (Zmax=7.53; θ=0) and we also showed that the phenotype segregated with either the K12 or the K3 locus in two Northern Irish pedigrees. Heterozygous missense mutations in K3 (E509K) and in K12 (V143L; R135T) completely co-segregated with MCD in the families and were not found in 100 normal unrelated chromosomes. All mutations occur in the highly conserved keratin helix boundary motifs, where dominant mutations in other keratins have been found to severely compromise cytoskeletal function, leading to keratinocyte fragility phenotypes. Our results demonstrate for the first time the molecular basis of Meesmann's corneal dystrophy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Moll, R., Franke, W.W., Schiller, D.L., Geiger, B. & Krepler, R. The catalog of human cytokeratins: Patterns of expression in normal epithelia, tumors and cultured cells. Cell 31, 11–24 (1982).
Sun, T.-T. et al. in The Transformed Phenotype (eds Levine, A.J., Vande Woude, G.F., Topp, W.C & Watson, J.D.) 169–176 (Cold Spring Harbor Laboratory, New York, 1984).
Meesmann, A. & Wilke, F. Klinische und anatomische Untersuchungen ueber eine bisher unbekannte, dominant vererbte Epitheldystrophie der Hornhaut. Klin. Mbl. Augenheilk. 103, 361–391 (1939).
Fine, B.S., Yanoff, M., Pitts, E. & Slaughter, F.D. Meesmann's epithelial dystrophy of the cornea. Am. J. Opthal. 83, 633–642 (1977).
Liu, C.Y. et al. Cornea-specific expression of K12 keratin during mouse development. Curr. Eye Res. 12, 963–74 (1993).
Lane, E.B. in Connective Tissue and its Heritable Disorders. Molecular, Genetic and Medical Aspects(eds. Royce, P.M. & Steinmann, B.) 237–247 (Wiley-Liss Inc., New York, 1993).
Quinlan, R.A., Hutchison, C.J. & Lane, E.B. Intermediate Filaments(Academic Press Ltd., London 1994).
McLean, W.H.I. & Lane, E.B. Intermediate filaments in disease. Curr. Opin. Cell Biol. 7, 118–125 (1995).
Corden, L.D. & McLean, W.H.I. Human keratin diseases: Hereditary fragility of specific epithelial tissues. Exp. Dermatol. 5, 297–307 (1996).
Schermer, A., Galvin, S. & Sun, T.-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell. Biol. 103, 49–62 (1986).
Rodrigues, M., Ben-Zvi, A., Krachmer, A. & Sun, T.-T. Suprabasal expression of a 65-kDa keratin (No.3) in developing human corneal epithelium. Differentiation 34, 60–67 (1987).
Chaloin-Dufau, C., Sun, T.-T. & Dhouailly, D. Appearance of the keratin pair K3/K12 during embryonic and adult corneal epithelial differentiation in the chick and in the rabbit. Cell Diff. Dev. 32, 97–108 (1990).
McKusick, V.A. OMIM: On-line Mendelian Inheritance in Man.(Johns Hopkins University, Baltimore, 1997).
Shearman, A.M. et al. The gene for Schnyder's crystalline corneal dystrophy maps to human chromosome 1p34.1-p36. Hum. Mol. Genet. 5, 1667–1672 (1996).
Stone, E.M. et al. Three autosomal dominant corneal dystrophies map to chromosome 5q. Nature Genet. 6, 47–51 (1994).
Small, K.W. et al. Mapping of Reis-Bucklers' corneal dystrophy to chromosome 5q. Am. J. Ophthal. 121, 384–390 (1996).
Vance, J. et al. Linkage of a gene for macular corneal dystrophy to chromosome 16. Am. J. Hum. Genet. 58, 757–762 (1996).
Toma, N.M. et al. Linkage of congenital hereditary endothelial dystrophy to chromosome 20. Hum. Mol. Genet. 4, 2395–2398 (1995).
Heon, E. et al. Linkage of posterior polymorphous corneal dystrophy to 20q11. Hum. Mol. Genet. 4, 485–488 (1995).
Skretting, G. & Prydz, H. An amino acid exchange in exon I of the human lecithin:cholesterol acyltransferase (LCAT) gene is associated with fish eye disease. Biochem. Biophys. Res. Commun. 182, 583–587 (1992).
Levy, E., Haltia, M. & Frangione, B. Amyloidosis due to a mutation of the gelsolin gene in an American family with lattice corneal dystrophy type II. N. Eng. J. Med. 325, 1780–1785 (1991).
Munier, F.L. et al. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nature Genet. 15, 247–251 (1997).
Tremblay, M. & Dube, I. Meesmann's corneal dystrophy: Ultrastructural features. Can. J. Opthalmol. 17, 24–28 (1982).
Ishida-Yamamoto, A. et al. Epidermolysis bullosa simplex (Dowling- Meara type) is a genetic disease characterized by an abnormal keratin filament network involving keratins K5 and K14J. J invest.Dermatol. 97, 959–968 (1991).
Kao, W.W.-Y et al. Keratin 12 deficient mice have fragile corneal epithelia. Ophthalmol. Vis. Sci. 37, 2572–2584 (1996).
Klinge, E.M., Sylvestre, Y.R., Freedberg, I.M. & Blumenberg, M. Evolution of keratin genes: Different protein domains evolve by different pathways. J. Mol. Evol. 24, 319–329 (1987).
Nishida, K. et al. A gene expression profile of human corneal epithelium and the isolation of human keratin 12 cDNA. Invest. Opthalmol. Vis. Sci. 37, 1800–1809 (1996).
Christiano, A.M. & Uitto, J. (1996) Molecular complexity of the cultaneous basement membrane zone. Revelations from the paradigm of epidermolysis bullosa. Exp. Dermatol. 5, 1–11.
McLean, W.H. et al.Keratin-16 and keratin-17 mutations cause pachyonychia-congenita. Nature Genet. 9, 273–278 (1995).
Kao, W.W.-Y Characterization and chromosomal localization of the cornea-specific murine keratin gene: Krt.12. J. Biol. Chem. 269, 24627–24636 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Irvine, A., Corden, L., Swensson, O. et al. Mutations in cornea-specific keratin K3 or K12 genes cause Meesmann's corneal dystrophy. Nat Genet 16, 184–187 (1997). https://doi.org/10.1038/ng0697-184
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0697-184
This article is cited by
-
The application and progression of CRISPR/Cas9 technology in ophthalmological diseases
Eye (2023)
-
Corneal dystrophies
Nature Reviews Disease Primers (2020)
-
In vivo histology and p.L132V mutation in KRT12 gene in Japanese patients with Meesmann corneal dystrophy
Japanese Journal of Ophthalmology (2019)
-
Seltene Erkrankungen in Deutschland und in Europa
Zeitschrift für Epileptologie (2019)
-
Comparative proteomic analysis of amnion membrane transplantation and cross-linking treatments in an experimental alkali injury model
International Ophthalmology (2018)