Abstract
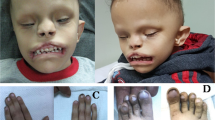
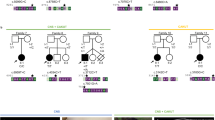
Holoprosencephaly (HPE) is the most common structural defect of the developing forebrain in humans (1 in 250 conceptuses, 1 in 16,000 live-born infants1,2,3). HPE is aetiologically heterogeneous, with both environmental and genetic causes4,5. So far, three human HPE genes are known: SHH at chromosome region 7q36 (ref. 6); ZIC2 at 13q32 (ref. 7); and SIX3 at 2p21 (ref. 8). In animal models, genes in the Nodal signalling pathway, such as those mutated in the zebrafish mutants cyclops (refs 9,10), squint (ref. 11) and one-eyed pinhead (oep; ref. 12), cause HPE. Mice heterozygous for null alleles of both Nodal and Smad2 have cyclopia13. Here we describe the involvement of the TG-interacting factor (TGIF), a homeodomain protein, in human HPE. We mapped TGIF to the HPE minimal critical region in 18p11.3. Heterozygous mutations in individuals with HPE affect the transcriptional repression domain of TGIF, the DNA-binding domain or the domain that interacts with SMAD2. (The latter is an effector in the signalling pathway of the neural axis developmental factor NODAL, a member of the transforming growth factor-β (TGF-β) family.) Several of these mutations cause a loss of TGIF function. Thus, TGIF links the NODAL signalling pathway to the bifurcation of the human forebrain and the establishment of ventral midline structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Muenke, M. & Beachy, P.A. Holoprosencephaly. in The Metabolic and Molecular Bases of Inherited Disease (eds Scriver, C.R. et al.) (McGraw-Hill, New York, in press)
Golden, J.A. Holoprosencephaly: a defect in brain patterning. J. Neuropathol. Exp. Neurol. 57, 991–999 (1998).
Ming, J.E. & Muenke, M. Holoprosencephaly: from Homer to Hedgehog. Clin. Genet. 53, 155–163 (1998).
Barr, M. Jr et al. Holoprosencephaly in infants of diabetic mothers. J. Pediatr. 102, 565–568 (1983).
Roessler, E. & Muenke, M. Holoprosencephaly: a paradigm for the complex genetics of brain development. J. Inherit. Metab. Dis. 21, 481–497 (1998).
Roessler, E. et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nature Genet. 14, 357–360 (1996).
Brown, S.A. et al. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nature Genet. 20, 180–183 (1998).
Wallis, D.E. et al. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nature Genet. 22, 196–199 (1999).
Sampath, K. et al. Induction of the zebrafish ventral brain and floorplate requires cyclops/Nodal signalling. Nature 395, 185–189 (1998).
Rebaglianti, M.R., Toyama, R., Haffter, P. & Dawid, I. Cyclops encodes a Nodal-related factor involved in midline signaling. Proc. Natl Acad. Sci. USA 95, 9932–9937 (1998).
Feldman, B. et al. Zebrafish organizer development and germ-layer formation require Nodal-related signals. Nature 395, 181–185 (1998).
Gritsman, K. et al. The EGF-CFC protein one-eyed pinhead is essential for Nodal signaling. Cell 97, 121–132 (1999).
Nomura, M. & Li, E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393, 786–790 (1998).
Bertolino, E., Reimund, B., Wildt-Perinic, D. & Clerc, R.G. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270, 31178–31188 (1995).
Edwards, M.C. et al. Human CPR (cell cycle progression restoration) genes impart a Far− phenotype on yeast cells. Genetics 147, 1063–1076 (1997).
Wotton, D., Lo, R.S., Lee, S. & Massagué, J. A Smad transcriptional corepressor. Cell 97, 29–39 (1999).
Massagué, J. TGFβ signal transduction. Annu. Rev. Biochem. 67, 753–791 (1998).
Luo, K. et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev. 13, 2196–2206 (1999).
Overhauser, J. et al. Physical mapping of the holoprosencephaly critical region in 18p11.3. Am. J. Hum. Genet. 57, 1080–1085 (1995).
Bertolino, E., Wildt, S., Richards, G. & Clerc, R.G. Expression of a novel murine homeobox gene in the developing cerebellar external granular layer during its proliferation. Dev. Dyn. 205, 410–420 (1996).
Wotton, D., Lo, R.S., Swaby L.A.C. & Massagué, J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J. Biol. Chem. 274, 37105–37110 (1999).
Liu, F., Massagué, J. & Ruiz i Altaba, A. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nature Genet. 20, 325–326 (1998).
Varlet, I. & Robertson, E.J. Left-right asymmetry in vertebrates. Curr. Opin. Genet. Dev. 7, 518–523 (1997).
Nanni, L. et al. The mutational spectrum of the Sonic Hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum. Mol. Genet. 13, 2479–2488 (1999).
Acknowledgements
We thank the patients and their families for participation; D. Mowat for clinical information; and K. Mahon and P. Beachy for discussions. K.W.G. was supported by the Howard Hughes Medical Institute. M.C.E. was supported by a Cellular and Molecular Biology of Aging Training Grant. This work was supported by NIH grants to M.M., J.M. and S.J.E.; by an NIH grant to the Memorial Sloan-Kettering Cancer Center; and by the Division of Intramural Research, National Human Genome Research Institute, NIH to M.M. S.J.E. and J.M. are Investigators with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gripp, K., Wotton, D., Edwards, M. et al. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet 25, 205–208 (2000). https://doi.org/10.1038/76074
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/76074
This article is cited by
-
Satb2 acts as a gatekeeper for major developmental transitions during early vertebrate embryogenesis
Nature Communications (2021)
-
Mosaic chromosome 18 anomaly delineated in a child with dysmorphism using a three-pronged cytogenetic techniques approach: a case report
BMC Medical Genomics (2020)
-
An enigmatic translocation of the vertebrate primordial eye field
BMC Evolutionary Biology (2020)
-
Genetics of atrioventricular canal defects
Italian Journal of Pediatrics (2020)
-
Genetic and Molecular Analyses indicate independent effects of TGIFs on Nodal and Gli3 in neural tube patterning
European Journal of Human Genetics (2017)