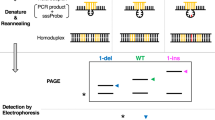
Abstract
Variations, such as nucleotide substitutions, deletions and insertions, within genes can affect the function of the gene product and in some cases be deleterious. Screening for known allelic variation is important for determining disease and gene associations1. Techniques which target specific mutations such as restriction enzyme polymorphism and oligonucleotide probe or PCR primer reactivity are useful for the detection of specific mutations, but these techniques are not generally effective for the identification of new mutations. Approaches for measuring changes in DNA conformation have been developed, based on the principle that DNA fragments which differ in nucleotide composition exhibit different mobilities after separation by polyacrylamide gel electrophoresis (PAGE; refs 2,3). Here we describe a conformation-based mutation detection system, double-strand conformation analysis (DSCA), which provides a simple means to detect genetic variants and to type complex polymorphic loci. We demonstrate the application of DSCA to detect genetic polymorphisms such as a single-nucleotide difference within DNA fragments of up to 979 base pairs in length. We present the application of DSCA in detecting four different mutations in the cystic fibrosis gene (CFTR) and 131 different alleles encoded by HLA class I genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Dykes, C., Genes, disease and medicine, Br. J. Clin. Pharmacol. 42, 683–695 (1996).
Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K. & Sekiya, T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphism. Proc. Natl. Acad. Sci. USA 86, 2766–2770 (1989).
Myers, R.M., Lumelsky, N., Lerman, L.S. & Maniatis, T. Detection of single base substitutions in total genomic DNA. Nature 313, 495–497 (1985).
Zimmerman, P., Carrington, M. & Nutman, T. Exploiting structural differences among heteroduplex molecules to simplify genotyping the DQA1 and DQB1 alleles in human lymphocyte typing. Nucleic Acids Res. 21, 4541–4547 (1993).
Arguello, R. et al. Complementary strand analysis: a new approach for allelic separation in complex polyallelic genetic systems. Nucleic Acids Res. 25, 2236–2238 (1997).
Verlingue, C. et al. Retrospective study of the cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in Guthrie cards from a large cohort of neonatal screening for cystic fibrosis. Hum. Genet. 93, 429–434 (1994).
Blasczyk, R., Hahn, U., Wehling, J., Huhn, D. & Salama, A. Complete subtyping of the HLA-A locus by sequence-specific amplification followed by direct sequencing or single-strand conformation polymorphism analysis. Tissue Antigens 46, 86–95 (1995).
Savage, D.A. et al. A rapid HLA-DRB1*04 subtyping method using PCR and DNA heterduplex generators. Tissue Antigens 47, 284–292 (1996).
Cereb, N., Maye, P., Lee, S., Kong, Y. & Yang, S.Y. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, And -C alleles. Tissue Antigens 45, 1–11 (1995).
Marsh, S.G.E. et al. The 12th International Histocompatibility Workshop cell lines panel, in Proceedings of the Twelfth International Histocompatibility Workshop and Conference Vol. I(ed. Charron, D.) 26–28 (EDK, Paris, 1997).
Lau, M., Terasaki, P. & Park, M., International Cell Exchange, (eds Terasaki, P.I. & Cecka, J.M.) 467–488 (UCLA Tissue-Typing Laboratory, Los Angeles, 1994)
Zielenski, J. et al. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 10, 214–228 (1991).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Argüello, J., Little, AM., Pay, A. et al. Mutation detection and typing of polymorphic loci through double-strand conformation analysis. Nat Genet 18, 192–194 (1998). https://doi.org/10.1038/ng0298-192
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0298-192