Abstract
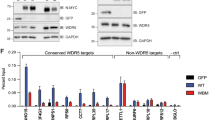
Myc and Mad family proteins regulate multiple biological processes through their capacity to influence gene expression directly. Here we show that the basic regions of Myc and Mad proteins are not functionally equivalent in oncogenesis, have separable E-box–binding activities and engage both common and distinct gene targets. Our data support the view that the opposing biological actions of Myc and Mxi1 extend beyond reciprocal regulation of common gene targets. Identification of differentially regulated gene targets provides a framework for understanding the mechanism through which the Myc superfamily governs the growth, proliferation and survival of normal and neoplastic cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Evan, G. & Littlewood, T.D. The role of c-myc in cell growth. Curr. Opin. Genet. Dev. 3, 44– 49 (1993).
Amati, B. & Land, H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 4, 102–108 (1994).
Blackwood, E.M., Kretzner, L. & Eisenman, R.N. Myc and Max function as a nucleoprotein complex. Curr. Opin. Genet. Dev. 2, 227–235 (1992).
Henriksson, M. & Luscher, B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68, 109–182 ( 1996).
Schreiber-Agus, N. & DePinho, R.A. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays 20, 808–818 (1998).
Blackwell, T.K., Kretzner, L., Blackwood, E.M., Eisenman, R.N. & Weintraub, H. Sequence-specific DNA binding by the c-Myc protein. Science 250, 1149– 1151 (1990).
Kretzner, L., Blackwood, E.M. & Eisenman, R.N. Myc and Max proteins possess distinct transcriptional activities. Nature 359, 426– 429 (1992).
Ayer, D.E., Kretzner, L. & Eisenman, R.N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72, 211 –222 (1993).
Ferre d'Amare, A., Prendergast, G.C., Ziff, E.B. & Burley, S.K. Recognition of Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45 ( 1993).
Grandori, C., Mac, J., Siebelt, F., Ayer, D.E. & Eisenman, R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 15, 4344–4357 (1996).
Solomon, D.L., Amati, B. & Land, H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 21, 5372–5376 (1993).
Halazonetis, T.D. & Kandil, A.N. Determination of the c-MYC DNA-binding site. Proc. Natl Acad. Sci. USA 88, 6162–6166 (1991).
Fisher, F. & Goding, C.R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 11, 4103– 4109 (1992).
Hurlin, P.J., Queva, C. & Eisenman, R.N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 11, 44–58 (1997).
Greenberg, R.A. et al. Telomerase reverse transcriptase is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18, 1219–1226 ( 1999).
Krikos, A., Laherty, C.D. & Dixit, V.M. Transcriptional activation and the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J. Biol. Chem. 267, 17971– 17976 (1992).
Chiorini, J.A., Miyamoto, S., Harkin, S.J. & Safer, B. Genomic cloning and characterization of the human eukaryotic initiation factor-2ß promoter. J. Biol. Chem. 274, 4195– 4201 (1999).
Dang, C.V. c-Myc target genes involved in cell growth, apoptosis and metabolism. Mol. Cell. Biol. 19, 1–11 (1999).
Grandori, C. & Eisenman, R.N. Myc target genes. Trends Biochem. Sci. 22, 177–181 (1997).
Felsher, D.W. & Bishop, J.M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl Acad. Sci. USA 96, 3940–3944 ( 1999).
Lee, T.C., Li, L., Philipson, L. & Ziff, E.B. Myc represses transcription of the growth arrest gene gas1. Proc. Natl Acad. Sci. USA 94, 12886–12891 (1997).
Marhin, W.M., Chen, S., Facchini, L.M., Fornace, A.J. Jr & Penn, L.Z. Myc represses the growth arrest gene gadd45. Oncogene 14, 2825– 2834 (1997).
Wu, K.J., Polack, A. & Dalla-Favera, R. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-Myc. Science 283, 676– 679 (1999).
Li, L.H., Nerlov, C., Prendergast, G., MacGregor, D. & Ziff, E.B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13, 4070–4079 (1994).
Lee, L.A. & Dang, C.V. c-Myc transrepression and cell transformation. Curr. Top. Microbiol. Immunol. 224, 131 –135 (1999).
Kauffmann-Zeh, A. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature 385, 544–548 (1997).
Mukherjee, B., Morgenbesser, S.D. & DePinho, R.A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 6, 1480–1492 (1992).
Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K. & Pease, L.R. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Schreiber-Agus, N. et al. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80, 777–786 (1995).
Jones, T.A., Zou, J.Y. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 ( 1991).
Ferre-D'Amare, A.R., Pognonec, P., Roeder, R.G. & Burley, S.K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189 (1994).
Shimizu, T. et al. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 16, 4689– 4697 (1997).
Christopher, J.A. SPOCK: The Structural Properties Observation and Calculation Kit (Texas A & M University, The Center for Macromolecular Design, College Station, 1998).
Serrano, M., Lin, A.W., McCurrach, M.E., Beach, D. & Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 ( 1997).
DeRisi, J. et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet. 14, 457– 460 (1996).
Khan, J. et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 58, 5009– 5013 (1998).
Chen, Y., Dougherty, E.R. & Bittner, M.L. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Optics 2, 364–374 (1997).
Acknowledgements
We thank H. Land for the CAT reporter constructs; S. Lowe for the mouse ecotropic receptor; S. Hann for the pBabe-c-MycER plasmid; J. Chiorini for the eIF2ß reporter; Y. Jiang and J.K. Lee for technical assistance; and members of the DePinho laboratory for helpful comments. R.C.O. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. N.S.-A. is a recipient of a Special Fellowship from the Leukemia Society of America. R.A.D. is supported by grants (RO1HD28317, RO1EY09300) from the National Institutes of Health and is an American Cancer Society Research Professor. Support from the DFCI Cancer Core grant to R.A.D. is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
O'Hagan, R., Schreiber-Agus, N., Chen, K. et al. Gene-target recognition among members of the Myc superfamily and implications for oncogenesis. Nat Genet 24, 113–119 (2000). https://doi.org/10.1038/72761
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/72761
This article is cited by
-
Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis
Nature Communications (2018)
-
PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci
BMC Bioinformatics (2010)
-
The interplay between MYC and HIF in cancer
Nature Reviews Cancer (2008)
-
The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein
Nature Cell Biology (2008)
-
Novel c‐MYC target genes mediate differential effects on cell proliferation and migration
EMBO reports (2007)